Abstract
Sarcophaga gracilior Chen, 1975 (Diptera: Sarcophagidae) plays a significant role in epidemiology and medicine. In this study, we first report the complete mitochondrial genome (mitogenome) of S. gracilior. This mitogenome was 15,534 bp in length (GenBank No. MW531675), comprising 13 protein-coding genes (PCGs), two ribosomal RNAs (rRNAs), 22 transfer RNAs (tRNAs), and a non-coding control region. The arrangement of genes was identical to that of ancestral metazoan. Nucleotide composition revealed a strong A + T bias, accounting for 76.7% (A 39.6%, G 9.3%, C 14.0%, T 37.1%). The phylogenetic relationships indicated that the species of S. gracilior emerged as sister to Sarcophaga melanura. This study provides important mitochondrial data for further studying evolutionary relationships and species identification of flesh flies.
The Sarcophagidae family plays a significant role in epidemiology and medicine (Ren et al. Citation2018). Sarcophaga gracilior Chen, 1975 (Diptera: Sarcophagidae), as known as Tricholioproctia (Hamimembrana) gracilior, has been recorded only in China and Nepal (Pape Citation1996). Here, we present the complete mitochondrial genome (mitogenome) of S. gracilior, which would further enrich our understanding of the phylogenetic relationship of Sarcophaga genus (Cameron Citation2014).
In this study, adult specimens were captured by pig livers in 4 August 2020, Guiyang city (25°46′N, 112°43′E), Guizhou province, China. All specimens were identified by traditional morphological keys, and then these specimens were stored at −80 °C in Guo’s lab (Hunan, Changsha, China) with a unique code (CSU2021012101). According to the manufacture's instruction, total DNA was extracted from thoracic muscle tissues of an adult specimen using QIANamp Micro DNA Kit (Qiangen Biotech Co., Ltd., Beijing, China). The mitogenome sequencing of S. gracilior was performed on an Illumina HiSeq 2500 Platform (Illumina, San Diego, CA), and then de novo assembly was carried out with MITObim v 1.9 and SOAPdenovo v2.04. Finally, the rough boundaries of all genes were initially identified by the MITOS2 Web Server (http://mitos2.bioinf.uni-leipzig.de/index.py) (Bernt et al. Citation2013).
In this study, the mitogenome of S. gracilior was first sequenced, which was 15,534 bp in length (GenBank No. MW531675), containing 13 protein-coding genes (PCGs), two ribosomal RNAs (rRNAs), 22 transfer RNAs (tRNAs), and a non-coding control region. The arrangement of genes was identical to that of ancestral metazoan (Cameron 2014). Nucleotide composition revealed a highly A + T bias, accounting for 76.7% (A 39.6%, G 9.3%, C 14.0%, T 37.1%).
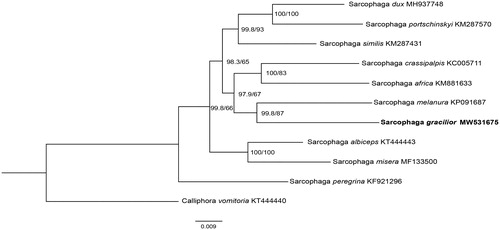
Phylogenetic analysis of S. gracilior with nine sarcophagids species was constructed using the maximum likelihood (ML) method based on the 13 PCGs, and Calliphora vomitoria (Diptera: Calliphoridae) was used as an outgroup (). ML analysis was performed by IQ-TREE v1.6.12 (Ren et al. Citation2020). In the phylogenetic tree, the species-level relationship between S. gracilior and Sarcophaga melanura closely clustered together, and separated clearly from the rest of species. The branches were well supported. This study provides significant mitochondrial data for further studying on evolutionary relationships and species identification of flesh flies.
Acknowledgment
The authors are grateful to Prof. Lushi Chen (Guizhou Police Officer College) for species identification.
Disclosure statement
The authors have declared that no competing interests exist.
Data availability statement
The data that support the findings of this study are openly available in NCBI at https://www.ncbi.nlm.nih.gov (GenBank: MW531675, BioProject: PRJNA695801, BioSample: SAMN17676972, SRA: SRR13608376). The sample was stored in Guo’s laboratory (Yadong Guo PhD, [email protected]) with a unique code CSU2021012101.
Additional information
Funding
References
- Bernt M, Donath A, Juhling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Cameron SL. 2014. Insect mitochondrial genomics: implications for evolution and phylogeny. Annu Rev Entomol. 59:95–117.
- Pape T. 1996. Catalogue of the Sarcophagidae of the World (Insecta: Diptera). Memoirs Entomol Int. 8:1–558.
- Ren L, Shang Y, Chen W, Meng F, Cai J, Zhu G, Chen L, Wang Y, Deng J, Guo Y, et al. 2018. A brief review of forensically important flesh flies (Diptera: Sarcophagidae). Forensic Sci Res. 3(1):16–26.
- Ren L, Zhang X, Li Y, Shang Y, Chen S, Wang S, Qu Y, Cai J, Guo Y. 2020. Comparative analysis of mitochondrial genomes among the subfamily Sarcophaginae (Diptera: Sarcophagidae) and phylogenetic implications. Int J Biol Macromol. 161:214–222.