Abstract
Eurasian Tree Sparrow Passer montanus is widely distributed passerine bird, and one sub-species P. m. saturatus is known to inhabit North-East Asia. In this study, we decode the complete mitochondrial genome of P. m. saturatus from the Republic of Korea. Mitogenome was 16,904 bp in length, and the content of A, T, G, and C were 30.0% (5079 bp), 22.5% (3810 bp), 15.5% (2621 bp), and 31.9% (5394 bp), respectively. The circular mitogenome contained 38 genes (13 protein-coding genes, 22 transfer RNAs, and 2 ribosomal RNAs) and a non-coding region. Phylogenetic analysis based on the complete mitogenome sequences indicated genetic distances in the species of Passeriformes, and P. m. saturatus in the Republic of Korea is included in a monophyletic group with P. montanus in China. This result provide basic information of population genetics of wide-ranging species Eurasian Tree Sparrow.
Keywords:
Introduction
Eurasian Tree Sparrow, Passer montanus, is a small passerine bird (family Passeridae) and is a very wide-ranging species. Its native range is distributed from Western Europe to Central Asia, East Asia, Indo-Chinese countries, and the Philippines, and was introduced to United States (Summer-Smith Citation1995). This species is reported to extant eleven geographic subspecies, of which three subspecies are distributed in the Northeast Asia: P. m. montanus (Linnaeus, 1758; in NE Mongolia, NE China, Russia), P. m. dilutus (Richmond, 1896; NW China SW Mongolia), and P. m. saturatus (Stejneger 1885; Summer-Smith Citation1995; Del Hoyo et al. Citation2018; South Korea, Taiwan, Japan, Sakhalin). Genetic research on complete mitochondrial genome (mitogenome) sequence of the P. m. saturatus was conducted at Anhui, China (Yang et al. Citation2016), but it has not been done in the Republic of Korea. In this study, we determined the whole mitogenome sequence of the P. m. saturatus and conducted phylogenetic analysis with related species. This study will help determining the phylogenetic position and evolution of the genome of this species.
To get the biological sample, a male individual P. m. saturatus (Ring color: blue green & sky) was captured from the Agricultural Practical Training Center (N 35°10'31.02", E 126°53'56.33", altitude: 40 m) at Chonnam National University, using mist net on 26 February 2020. We collected blood sample from capillary vessel by using Microsyringes, and stored the sample in EDTA vial (Voucher no. CNU2020-TSB140) at −20 °C, Research Center of Ecomimetics in Chonnam National University. The total genomic DNA was extracted by using the DNeasy Blood and Tissue kit (Qiagen, Valencia, CA) according to the manufacturer’s protocol, and the extracted DNA was stored at the −20 °C freezer. In this study, we sequenced the complete mitogenome of the P. m. saturatus by PCR-based approach (PRINZA 692774). We determined the complete mitogenome sequence using the direct sequencing (1000-bp length in each read) generated from Applied Biosystems 3730XL DNA Analyzer (Bionics Co., Seoul, Republic of Korea). Sequences arranged using de novo assembly and annotation by comparing P. montanus mitogenome (Accession no. JX486030) and P. m. saturatus mitogenome (Accession no. KM577704) in GenBank, and analysis mitogenome structure using Geneious prime 2021.0.3 (www.geneious.com). Phylogenetic analysis was constructed using maximum-likelihood method (Rzhetsky and Nei Citation1992) using MEGA X software with 100 replicates bootstrap method (Kumar et al. Citation2018).
The complete mitogenome of P. m. saturatus was 16,904 bp in length deposited in GenBank (Accession no. MW495245), and contains 13 protein-coding genes (PCGs), 22 transfer RNA (tRNA) genes, 2 ribosomal RNA genes (rRNA), and a putative long non-coding control region (NCR). The order and orientation are identical with that of the standard avian gene order, and other P. montanus’s mitogenome sequence (Gibb et al. Citation2006; Yang et al. Citation2016). The nucleotide composition of the P. m. saturatus in the Republic of Korea (A = 30.0%, T = 22.5%, G = 15.5%, C = 31.9%) was similar to that of P. montanus in China (JX486030.1; A = 30.0%, T = 22.5%, G = 15.5%, C = 31.9%), and P. m. saturatus in China (KM577704.1; A = 30.0%, T = 22.5%, G = 15.5%, C = 31.9%). Comparison of sequences between P. m. saturatus in the Republic of Korea and P. montanus. in China indicated a 99.62% sequence identity, and comparison with P. m. saturatus in China was 99.61% identity ().
Figure 1. Phylogenetic tree of Passer montanus saturatus in the Republic of Korea (underlined; MW495245) and other related species based on mitochondrial (mt) genome full sequence data. Streptopelia orientalis was used as an outgroup. In the phylogenetic tree, evolutionary distances were expressed as branch length. The evolutionary distances computed using the maximum likelihood method, and were in the units of the number of base substitutions per site. Bootstrap method used 100 replicates to know statistical support. The phylogenetic analysis was performed using MEGA X (Kumar et al. Citation2018).
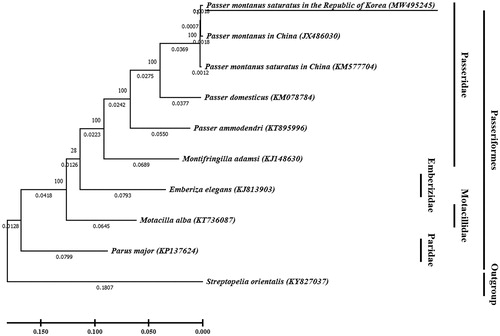
For phylogenetic analysis to investigate genetic relationship of P. montanus saturatus in Korea, the evolutionary history was inferred by using the maximum likelihood method with Tamura-Nei model (Tamura and Nei Citation1993). The full mitogenome sequences of nine Passeriformes were extracted from Genbank, with Streptopelia orientalis used as an outgroup. The phylogenetic tree showed that the close relationship between P. m. saturatus in the Republic of Korea and P. montanus in China (JX486030) than P. m. saturatus in China (KM577704). The phylogenetic relationship showed that the genetic distance increased as it progressed to the Passeridae, Emberizidae, Motacillidae, and Paridae in the Passeriformes. These data provide genetic information for P. m. saturatus resided in the Republic of Korea, for further studies about geographical species distribution of the Eurasian Tree Sparrow.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
GenBank accession number and BioProject Accession number from the complete mitochondrial genome of Passer montatus subsp. saturatus (GenBank Accession no. MW495245, BioProject Accession no. PRJNA692774) has been registered with the NCBI database. A specimen was deposited at Research Center of Ecomimetics in CNU under the sample number CNU2020-TSB140 (E-mail address of the person in charge: [email protected]).
Additional information
Funding
References
- Del Hoyo J, Elliott A, Sargatal J, Christie DA, de Juana EE. 2018. Handbook of the birds of the world alive. Barcelona: Lynx Edicions.
- Gibb GC, Kardailsky O, Kimball RT, Braun EL, Penny D. 2006. Mitochondrial genomes and avian phylogeny: complex characters and resolvability without explosive radiations. Mol Biol Evol. 24(1):269–280.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Rzhetsky A, Nei M. 1992. A simple method for estimating and testing minimum evolution trees. Mol Biol Evol. 9:945–967.
- Summer-Smith JD. 1995. The tree sparrow. Bath (UK): Bath Press.
- Tamura K, Nei M. 1993. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol. 10:512–526.
- Yang F, Li B, Zhou L, Bao D, Zhu H. 2016. Complete mitochondrial genome of Tree Sparrow Passer montanus saturatus (Passeriformes: Passeridae). Mitochondrial DNA Part A. 27(3):1993–1994.
