Abstract
Scutellaria meehanioides C.Y.Wu is a medicinal perennial herb native to China. The complete chloroplast genome sequence of the S. meehanioides was determined and assembled using next generation sequencing methodologies. The complete genome is 152,484 base pairs (bp) in length and has an overall GC content of 38.4%. The chloroplast genome contains, a large single-copy region (LSC) of 83,859 bp, small single-copy region (SSC) of 17,467 bp and a pair of inverted repeats (IRs) of 24,029 bp. The genome of S. meehanioides contains 133 genes, including 87 protein-coding, 38 tRNA,and 8 rRNA genes. Phylogenetic analysis fully resolved S. meehanioides in a clade with S. orthocalyx. This study provides useful information for future genetic study of S. meehanioides.
Scutellaria L. is species-rich, with more than 300 species currently accepted and widely distributed in Europe, the United States and East Asia (Zhang et al. Citation1998). Some species in Scutellaria have been used as local medicine for thousands years (Shang et al. Citation2010). Scutellaria meehanioides was one of them which is a perennial herb classified in the Lamiaceae and mainly distributed in Shaanxi Province, China . The root of S. meehanioides was used as a crude medicine for antitumor, anti-oxidant and hepato-protective (Yang et al. Citation2020). In this study, we report the complete chloroplast genome of S. meehanioides, which revealed the phylogenetic relationship with other species in Scutellaria and the Lamiaceae.
The samples of S. meehanioides were collected from Mt. Wutai, Xi'an City, Shaanxi Province (33°59′18.45″N, 108°58′23.44″E). The voucher specimen was deposited in XBGH (The Herbarium of Xi’an Botanical Garden, http://www.xazwy.com) (Voucher number: Xun Lulu et al.00918, Lulu Xun, [email protected]). The total genomic DNA was extracted from fresh leaves using the CTAB extraction method (Doyle and Doyle Citation1987). The genomic library was sequenced using the Illumina NovaSeq platform. Raw PE reads of about 2.4 Gb were trimmed and filtered, using fastp application (Chen et al. Citation2018), resulting in about 2.38 Gb clean PE reads. NOVOplasty v2.7.2 (Dierckxsens et al. Citation2016) was used for the de novo assembly of chloroplast genome using S. baicalensis (GenBank accession no. MF521663) as the seed reference and the parameters ‘Type = chloro K-mer = 39 Genome range = 13,000–17,000’. The sequence was annotated using GeSeq (https://chlorobox.mpimp-golm.mpg.de/geseq-app.html) and visually checked in Geneious v11.0.3 (Kearse et al. Citation2012) using the chloroplast genome of S. tsinyunensis (GenBank accession: NC 050761) as a reference. The complete chloroplast genome of S. meehanioides is publicly available under GenBank accession number MW381011.
The complete chloroplast genome of S. meehanioides is 152,484 bp in length and has an overall GC content of 38.4%, consisting of a small single-copy region (SSC) of 17,467 bp, a pair of inverted repeat regions (IRa and IRb) with the same length of 24,029 bp, and a large single-copy region (LSC) of 83,859 bp. One hundred thirty-three genes were predicted, including 87 protein-coding, 38 tRNA, and 4 rRNA genes.
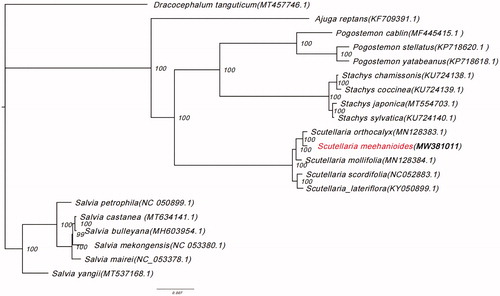
To confirm the phylogenetic position of S. meehanioides, the maximum likelihood (ML) phylogenetic analyses were conducted by IQTREE v1.6.7 (Nguyen et al. Citation2015) based on 20 complete chloroplast genome sequences of Lamiaceae, under TVM + F+R3 model with 5000 bootstrap replicates. The phylogenetic tree revealed that S. meehanioides was closely related to S. orthocalyx (). Some plastid genes, such as matK-trnK, atpB-atpE, psbC-psbD, and rps3-rpl22, overlap each other in the Lamiaceae chloroplast genomes (Liang et al. Citation2019). The newly characterized S. meehanioides complete chloroplast genome provides a reference for the phylogenetic relationships and assessment of the genetic structure of Scutellaria and the Lamiaceae.
Figure 1. Phylogenetic tree showing the relationship between Scutellaria meehanioides and 19 Lamiaceae species. Phylogenetic tree was constructed based on the complete chloroplast genomes using maximum likelihood (ML) with 5000 bootstrap replicates. Numbers in each node indicate the bootstrap support values.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI (https://www.ncbi.nlm.nih.gov) under the Accession no. MW381011. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA715065, SRR13987450, and SAMN18325271, respectively.
Additional information
Funding
References
- Chen S, Zhou Y, Chen Y, Gu J. 2018. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 34(17):i884–i890.
- Dierckxsens N, Mardulyn P, Smits G. 2016. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):1–9.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure from small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Liang C, Wang L, Lei J, Duan B, Ma W, Xiao S, Qi H, Wang Z, Liu Y, Shen X, et al. 2019. A comparative analysis of the chloroplast genomes of four Salvia medicinal plants. Engineering. 5(5):907–915.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Shang X, He X, He X, Li M, Zhang R, Fan P, Zhang Q, Jia Z. 2010. The genus Scutellaria an ethnopharmacological and phytochemical review. J Ethnopharmacol. 128(2):279–313.
- Yang Y, Tian J, Chen Z. 2020. Research progress on pharmacological action and chemical composition of Huangqin (RadixScutellariae) by processing. Chinese Arch Tradit Chinese Med. 10(38):2–4.
- Zhang YY, Don HY, Guo YZ, Ageta H, Harigaya Y, Onda M, Hashimoto K, Ikeya Y, Okada M, Maruno M, et al. 1998. Comparative study of Scutellaria planipes and Scutellaria baicalensis. Biomed Chromatogr. 12(1):31–33.
