Abstract
In this study, we sequenced and annotated the complete mitochondrial genome (mitogenome) of Neotoxoptera formosana (Takahashi) (Hemiptera: Aphididae). The complete mitogenome of N. formosana is 15,642 bp in length, and includes 13 protein-coding genes (PCGs), 2 ribosome RNA (rRNA) genes, 22 transfer RNA (tRNA) genes, and one control region. The overall base composition was as follows: 45.2% of A, 5.8% of G, 10.5% of C, and 38.4% of T, with a total of A + T content of 83.6%. The phylogenetic tree showed that N. formosana and Myzus persicae were clustered into one branch. This result will enrich the mitogenome of family Aphididae.
Onion aphid, Neotoxoptera formosana (Takahashi) (Hemiptera: Aphididae), originally occurs in Taiwan, China, and is reported in countries all over the world, such as Japan, Brazil, United Kingdom, Italy, Germany and Netherlands (Melo et al. Citation2005; Pellizzari and Montà Citation2005; Piron Citation2010). Onion aphid is an oligophagous aphid pest of Allium crops which rapidly builds up large colonies and damages the leaves (Hori Citation2007; Piron Citation2010). In the last year, we found that N. formosana is a main pest in Allium tuberosum in Guizhou Province, China (Wang et al. Citation2020). Previous studies mainly focused on ecological characteristic, taxonomic categories and partial sequences of N. formosana. In this study, we determined the mitogenome sequence of N. formosana to accelerate the population genetics and evolution of the genus Neotoxoptera.
The samples were collected from Puding County, Anshun City, Guizhou Province, China in Oct. 2020 (105°27′49ʺE, 26°26′36ʺN). Voucher specimens were deposited in the Institute of Entomology, Guizhou University, China (Jian-Feng Liu,[email protected]) and the label number was GUGC-Neo-00204. The total DNA was extracted from seven female adults of N. formosana by using the DNeasy Blood & Tissue kit (Cat. No. 69504). An Illumina ReSeq library was prepared with an average insert size of 400 bp and sequenced using the Illumina NovaSeq6000 platform with 150 bp paired-end (Berry Genomics, Beijing, China). The complete mitogenome sequence was assembled using NOVOplasty v2.7.2 (Dierckxsens et al. Citation2017) with K-mer value, and annotated by MitoZ v2.4 (Meng et al. Citation2019) with default set. Genomic annotation was calibrated using MITOS2 (Bernt et al. Citation2013) and Geneious Prime v2020.2.4 (Kearse et al. Citation2012).
The complete mitogenome of N. formosana was a circular double-stranded and 15,642 bp in length (GeneBank accession number MW534268), containing 37 encoding genes (13 PCGs, 22 tRNA genes, and 2 rRNA genes) and one control region. The whole base composition was as follows: 45.2% of A, 5.8% of G, 10.5% of C, and 38.4% of T, with a total of A + T content of 83.6%. All PCGs began with the typical ATN codons (ATT for COX1, ATP8, ATP6, ND3, ND5, ND6, and ND1; ATG for COX3, ND4, ND4L, and CYTB; ATA for ND2 and COX2). All protein coding genes used TAA as stop codon. The 16 s rRNA gene was 1,263 bp in size and was located between trnL and trnV, while the 12 s rRNA gene was 812 bp in size and was located behind trnV.
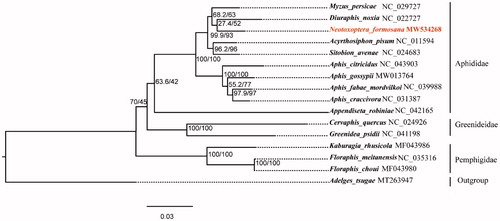
The nucleotide sequences of 13 PCGs (delete the third codon position) and 2 rRNA genes from 15 Aphidoidea species and one outgroup taxa from Pyrrhocoroidea species were aligned using MAFFT v7.394 with L-INS-I algorithm (Katoh and Standley Citation2013). The poorly aligned results were removed by trimAl v1.4.1 (Capella-Gutiérrez et al. Citation2009). A phylogenetic tree was constructed for 16 species by maximum likelihood method using IQ-TREE v1.6.3 software under the GTR + I + G model (Nguyen et al. Citation2015). The result confirmed that N. formosana and Myzus persicae were clustered into one branch ().
Disclosure statement
No potential conflict of interest was reported by the author(s).
Date availability statement
The data that support the findings of this study are openly available in [NCBI] at [https:www.ncbi.nlm.nih.gov/], reference number[MW534268]. The associated BioProject, BioSample, and SRA numbers are PRJNA703062, SAMN18011301, SRR13753265, respectively (https://www.ncbi.nlm.nih.gov/sra/?term=SRR13753265).
Additional information
Funding
References
- Bernt M, Donath A, Juhling F, Externbrink F, Florentz C, Fritzsch G, Putz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69:313–319.
- Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 25:1972–1973.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: denovo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Hori M. 2007. Onion aphid (Neotoxoptera formosana) attractants, in the headspace of Allium fistulosum and A. tuberosum leaves. J Appl Entomology. 131(1):8–12.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kearse M, Moir R, Wilson A, Stone-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markovitz S, Duran C, et al. 2012. Geneius basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Melo Filho PDA, Dusi AN, Costa CL, Resende RDO. 2005. Colonization of garlic plants by Neotoxoptera formosana in Distrito Federal, Brazil. Hortic Bras. 23(4):929–930.
- Meng GL, Li YY, Yang CT, Liu SL. 2019. Mitoz: a toolkit for animal mitochondrial genome assembly, annotation and visualization. Nucleic Acids Res. 47(11):e63.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 3(1):268–274.
- Pellizzari G, Montà LD, Vacante V. 2005. Alien insect and mite pests introduced to Italy in sixty years (1945–2004). Plant protection and plant health in Europe. Introduction and spread of invasive species. Berlin, Germany: Humbolt University; p. 9–11.
- Piron PGM. 2010. Appearance of Neotoxoptera formosana (Homoptera: Aphididae) in the Netherlands. Entomol Ber. 70(1):10–12.
- Wang T, Zhang H, Zeng C, Ren Y-L, Zhao Q-Q, Chen S-S, Tan X-F, Yang M-F, Liu J-F. 2020. New records of Rhizoglyphus robini (Acari: Acaridae) and Bradysia odoriphaga (Diptera: Sciaridae) infesting Allium tuberosum in Guizhou Province, China. Syst Appl Acarol. 25(12):2315–2319.