Abstract
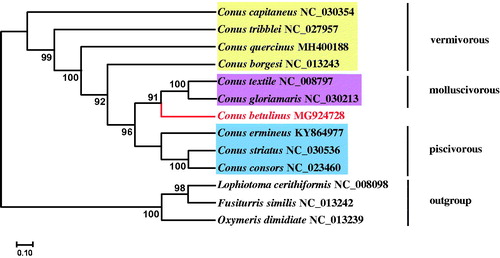
The complete mitochondrial genome of the tubular cone snail Conus betulinus is presented in this study. The C. betulinus mitochondrial genome was 16,240 bp with 13 protein-coding genes (PCGs), 22 transfer RNA (tRNA) genes, 2 ribosomal RNA (rRNA) genes, and a non-coding AT-rich region (D-loop). The overall base composition was estimated to be 25.67% for A, 38.26% for T, 21.38% for G, and 14.69% for C, with a high A + T content of 63.93%. Phylogenetic analyses based on 13 PCGs showed the close relationship of vermivorous C. betulinus with the common ancestor of molluscivorous Conus textile and Conus gloriamaris, providing a basis for further studies on the phylogenetics of cone snails according to their dietary type.
Members of the genus Conus (Neogastropoda: Conidae), poisonous carnivores that live in tropical ocean waters around the world, produce complex conotoxins for hunting and defense. More than 700 extant species and 57 subgenera are recognized in this classification (Puillandre et al. Citation2014; Gao, Peng, Chen, et al. Citation2018). As the largest genus among marine invertebrates, cone snails can be divided into the following three groups based on their diet: piscivorous, molluscivorous, and vermivorous (Gao et al. Citation2017). Vermivorous species account for 70% of the genus, and some studies have been conducted on the dominant vermivorous species that seem to be non-threatening (Himaya et al. Citation2015; Peng et al. Citation2016). In recent years, researchers have attempted to elucidate the evolutionary relationship between venom and feeding from population genetics, evolutionary biology, and phylogenetics (Aman et al. Citation2015; Gao, Peng, Chen, et al. Citation2018).
We assembled and characterized the complete mitochondrial genome sequence of vermivorous Conus betulinus Linnaeus 1758 (Neogastropoda: Conidae) to provide information for the identification of Conus and support further phylogenetic studies of Neogastropoda. Live C. betulinus was collected from the offshore areas of Sanya City, Hainan Province (18°09N, 108°56E). The sample was deposited at the Key Laboratory of Tropical Translational Medicine of the Ministry of Education, Hainan Medical University, Haikou, Hainan, China. The specimen accession number was CHMU0119. Total genomic DNA was extracted using the Column mtDNAout kit (Tianda, Beijing, China), and the purified genomic DNA was characterized using the Nanodrop 2000 spectrometer (Thermo Fisher Scientific, Wilmington, DE, USA). The NEBNext DNA Library Prep Kit (New England Biolabs, Ipswich, MA, USA) was used to establish a paired-end library. Quantification and sizing of the library were performed on a Bioanalyzer 2100 High Sensitivity DNA chip (Agilent, Palo Alto, CA, USA). The library was normalized to 2 nM and sequenced on the Illumina HiSeq2000 platform (Illumina, San Diego, CA, USA). The mitochondrial genome was assembled using SPAdes v.3.5.0 (Bankevich et al. Citation2012) with k-mer size 79, the average sequencing depth of mitochondrial genome is 85× and coverage 100% of the genome. Gene annotation was performed using the online programs Dual Organellar GenoMe Annotator (DOGMA) (Wyman et al. Citation2004), ORF Finder (Cheng et al. Citation2013), tRNAscan-SE (Schattner et al. Citation2005), and ARWEN with the annotated results of ARAGORN (Abe et al. Citation2011). All protein-coding genes (PCGs), transfer RNAs (tRNAs), ribosomal RNAs (rRNAs), and a non-coding AT-rich region (D-loop) were confirmed using Basic Local Alignment Search Tool (BLAST) (Altschul et al. Citation1990), tRNAscan-SE, and ARWEN (Griffiths-Jones et al. Citation2003; Abe et al. Citation2011).
The mitochondrial genome was a circular molecule of 16,240 bp in size with 13 PCGs, 22 tRNA genes, 2 rRNA genes, and a D-loop (GenBank accession number: MG924728). Eight tRNA genes were encoded on the light strand (tRNA-Met, tRNA-Tyr, tRNA-Cys, tRNA-Trp, tRNA-Gln, tRNA-Gly, tRNA-Glu, and tRNA-Thr), and the other 30 genes were located on the heavy strand. The overall base composition was estimated to be 25.67% for A, 38.26% for T, 21.38% for G, and 14.69% for C, with a high A + T content of 63.93%. The D-loop region between tRNA-Phe and cox3 in C. betulinus was the longest (793 bp) among the regions of the Conus species, which had a higher A + T content (69.86%). The genes order of Oxymeris dimidiate (NC_013239), Fusiturris similis (NC_013242), and C. betulinus are coincident, except for tRNA-Val and tRNA-Ser, and they were major rearrangement were found in Neogastropoda (Duda Citation2001; Xing and Lee Citation2005; Bandyopadhyay et al. Citation2008).
To investigate the phylogenetic position of C. betulinus within Conus, the 13 PCGs of mitogenomes from 9 Conus species and 3 allochthonous species were downloaded from GenBank, and phylogenetic trees were constructed using the maximum-likelihood (ML) method with RAxML (version 8.1.5) (Stamatakis Citation2006). Vermivorous cone snails were clustered at the root of the tree, implying the cone snail ancestor may be vermivorous (Gao, Peng, Zhu, et al. Citation2018). However, the vermivorous clade is not monophyletic but paraphyletic to the other two dietary types. It can be seen that vermivorous C. betulinus was closely related to the common ancestor of molluscivorous Conus textile and Conus gloriamaris (). The complete mitochondrial genome of the tubular cone snail C. betulinus will provide essential data for future research on the phylogenetic and evolutionary relationship in genus of Conus.
Disclosure statement
The authors declare no conflict of interest. The authors alone are responsible for the content and writing of this paper.
Data availability statement
The data that support the findings of this study are available in the NCBI database at https://www.ncbi.nlm.nih.gov, reference number MG924728.
Additional information
Funding
References
- Abe T, Ikemura T, Sugahara J, Kanai A, Ohara Y, Uehara H, Kinouchi M, Kanaya S, Yamada Y, Muto A, et al. 2011. tRNADB-CE 2011: tRNA gene database curated manually by experts. Nucleic Acids Res. 39:D210–D213.
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol. 215(3):403–410.
- Aman JW, Imperial JS, Ueberheide B, Zhang MM, Aguilar M, Taylor D, Watkins M, Yoshikami D, Showers-Corneli P, Safavi-Hemami H, et al. 2015. Insights into the origins of fish hunting in venomous cone snails from studies of Conus tessulatus. Proc Natl Acad Sci U S A. 112(16):5087–5092.
- Bandyopadhyay PK, Stevenson BJ, Ownby JP, Cady MT, Watkins M, Olivera BM. 2008. The mitochondrial genome of Conus textile, coxI-coxII intergenic sequences and Conoidean evolution. Mol Phylogenet Evol. 46(1):215–223.
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Cheng J, Zeng X, Ren G, Liu Z. 2013. CGAP: a new comprehensive platform for the comparative analysis of chloroplast genomes. BMC Bioinf. 14:95.
- Duda TF, Kohn AJ, Palumbi SR. 2001. Origins of diverse feeding ecologies within Conus, a genus of venomous marine gastropods. Biol J Linn Soc. 73(4):391–409.
- Gao B, Peng C, Chen Q, Zhang J, Shi Q. 2018. Mitochondrial genome sequencing of a vermivorous cone snail Conus quercinus supports the correlative analysis between phylogenetic relationships and dietary types of Conus species. PLOS One. 13(7):e0193053.
- Gao B, Peng C, Yang J, Yi Y, Zhang J, Shi Q. 2017. Cone snails: a big store of conotoxins for novel drug discovery. Toxins (Basel). 9(12):397.
- Gao B, Peng C, Zhu Y, Sun Y, Zhao T, Huang Y, Shi Q. 2018. High throughput identification of novel conotoxins from the vermivorous oak cone snail (Conus quercinus) by transcriptome sequencing. IJMS. 19(12):3901.
- Griffiths-Jones S, Bateman A, Marshall M, Khanna A, Eddy SR. 2003. Rfam: an RNA family database. Nucleic Acids Res. 31(1):439–441.
- Himaya SW, Jin AH, Dutertre S, Giacomotto J, Mohialdeen H, Vetter I, Alewood PF, Lewis RJ. 2015. Comparative venomics reveals the complex prey capture strategy of the piscivorous cone snail Conus catus. J Proteome Res. 14(10):4372–4381.
- Peng C, Yao G, Gao BM, Fan CX, Bian C, Wang J, Cao Y, Wen B, Zhu Y, Ruan Z, et al. 2016. High-throughput identification of novel conotoxins from the Chinese tubular cone snail (Conus betulinus) by multi-transcriptome sequencing. GigaScience. 5:17.
- Puillandre N, Bouchet P, Duda TF, Jr, Kauferstein S, Kohn AJ, Olivera BM, Watkins M, Meyer C. 2014. Molecular phylogeny and evolution of the cone snails (Gastropoda, Conoidea). Mol Phylogenet Evol. 78:290–303.
- Schattner P, Brooks AN, Lowe TM. 2005. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 33:W686–W689.
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 22(21):2688–2690.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20(17):3252–3255.
- Xing Y, Lee C. 2005. Assessing the application of Ka/Ks ratio test to alternatively spliced exons. Bioinformatics. 21(19):3701–3703.