Abstract
Lysmata vittata is widely distributed in the Indian and Pacific oceans. In this study, we sequenced the mitochondrial genome of L. vittata using Illumina HiSeq. The complete mitochondrial genome of L. vittata was 20,837 bp in length, including 13 protein-coding genes, 22 transfer RNA genes, and two ribosomal RNA genes. The contents of the four bases in the mitochondrial DNA were A (31.68%), T (32.36%), C (21.65%), and G (14.31%). Phylogenetic analysis of 41 shrimp showed that L. vittata was clustered with other Hippolytidae shrimp.
The red-striped shrimp Lysmata vittata (Decapoda: Hippolytidae) is a popular shrimp in the marine aquarium trade (Marin et al. Citation2012). It is widely distributed in the Indian and Pacific oceans, including along the coasts of Japan, China, Indonesia, Australia, and eastern Africa (Wang and Sha Citation2018; Alves et al. Citation2019). The species is commonly found at depths of 2 to 50 m and lives in large groups among rocks, algae, and sponges (Alves et al. Citation2019). Research on this species remains limited, although several studies have investigated its mating, larval development, gonadal development, and diet (Yang and Kim Citation2010; Chen et al. Citation2019). The complete mitochondrial genome (mitogenome) of L. vittata is an important resource for evolutionary research. In this study, we report on the complete mitogenome of L. vittata and analyze its phylogenetic relationships within Hippolytidae.
Specimens (voucher no. QP20200526-1) were collected from the South China Sea (22°36′ N, 114°32′ E), Shenzhen, Guangdong Province, China, and were stored in the herbarium of Qingyuan Polytechnic (Guangdong, China). Muscle samples of L. vittata were dissected and preserved at −80 °C until use. The muscle tissue was used for mitochondrial DNA (mtDNA) extraction with a TIANamp Marine Animals DNA Kit (Tiangen, Beijing, China) according to the manufacturer’s specifications. The mtDNA was sequenced using Illumina HiSeq (Illumina Inc., San Diego, CA, USA). Clean data were acquired and assembled using SPAdes v3.15.2 (Bankevich et al. Citation2012). MITO (http://mitos.bioinf.uni-leipzig.de/index.py) (Bernt et al. Citation2013) and ORF finder (https://www.ncbi.nlm.nih.gov/orffinder/) were used to identify and annotate protein-coding, transfer RNA (tRNA), and ribosomal RNA (rRNA) genes. Phylogenetic analysis was conducted using maximum-likelihood (ML) in MEGA X (Kumar et al. Citation2018).
The mitogenome of L. vittata was 20,837 bp in length (GenBank accession number: MW285083.1), which is larger than that of other species belonging to the Lysmata genus. This may be due to the 4-kb noncoding sequence between cox1 and cox2 in the L. vittata mitogenome, which is much longer than that of other species of Lysmata. The tRNA-Leu divided this noncoding sequence into two sequences.One was 1712 bp length with A + T content of 71.67%, and the other was 2270 bp length with A + T content of 72.82%. The L. vittata mitogenome contained 13 protein-coding, 22 tRNA, and two rRNA genes. Of the 37 genes, 22 were encoded by the heavy strand and 15 genes, including four protein-coding (ND1, ND4, ND4L, and ND5), two rRNA, and nine tRNA genes, were encoded by the light strand. The A, G, C, and T contents of the heavy strand were 34.36%, 11.74%, 17.03%, and 36.87%, respectively, with a high A + T content of 71.24%. All protein-coding genes had ATN as the start codon. Ten protein-coding genes (ND1, ND2, ND3, ND4L, ND6, Cox2, Cox3, ATP8, ATP6, and Cytb) contained a TAA stop codon, one protein-coding gene (Cox1) contained a TAG stop codon, and two protein-coding genes (ND4 and ND5) contained an incomplete T– stop codon. 16S rRNA and 12S rRNA were 1 467 bp (72.26% AT content) and 821 bp (67.07% AT content) in length, respectively. All tRNA genes ranged from 51 to 77 bp in size.
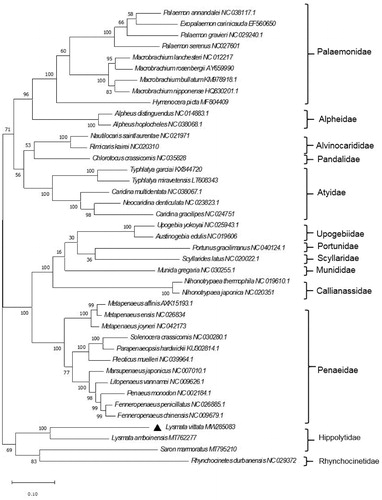
Based on the 13 complete concatenated protein-coding genes of 41 shrimp from the GenBank database, a phylogenetic tree was constructed using the maximum-likelihood (ML) method and Jones-Taylor-Thornton (JTT) matrix-based model, with a bootstrap of 1 000 replicates (). The phylogenetic tree showed that L. vittata was clustered with L. amboinensis. However, Saron marmoratus and Rhynchocinetes durbanensis were clustered together. This result was similar to the results of previous research (Terossi et al. Citation2017; Wang et al. Citation2021), which all proved that Hippolytidae can be considered as a polyphyletic taxon. In conclusion, our study described the complete mitogenome of L. vittata and analyzed its phylogenetic position within Hippolytidae. This research should contribute to further investigations on the molecular evolution and conservation of this species.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in NCBI at https://www.ncbi.nlm.nih.gov/, reference number MW285083.
Additional information
Funding
References
- Alves DFR, López GL, Barros-Alves SP, Hirose GL. 2019. Sexual system, reproductive cycle and embryonic development of the red-striped shrimp Lysmata vittata, an invader in the western Atlantic Ocean. PLoS One. 14(1):e0210723.
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Chen D, Liu F, Zhu Z, Lin Q, Zeng C, Ye H. 2019. Ontogenetic development of gonads and external sexual characters of the protandric simultaneous hermaphrodite peppermint shrimp, Lysmata vittata (Caridea: Hippolytidae). PLoS One. 14(4):e0215406.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Marin IN, Korn OM, Kornienko ES. 2012. The caridean shrimp Lysmata vittata (Stimpson, 1860) (Decapoda: Hippolytidae): a new species for the fauna of Russia. Russ J Mar Biol. 38(4):359–363.
- Terossi M, De Grave S, Mantelatto FL. 2017. Global biogeography, cryptic species and systematic issues in the shrimp genus Hippolyte Leach, 1814 (Decapoda: Caridea: Hippolytidae) by multimarker analyses. Sci Rep. 7(1):6697.
- Wang Y, Zeng L, Wen J, Li X, Huang Y, Sun Y, Zhao J. 2021. Characterization of the complete mitochondrial genome of Saron marmoratus (Hippolytidae, Decapoda) and its phylogenetic analysis. Mitochondrial DNA B Resour. 6(1):124–126.
- Wang YR, Sha ZL. 2018. Description of two new species of Lysmata Risso, 1816 (Decapoda, Lysmatidae) from the China seas, with remarks on Lysmata vittata (Stimpson 1860). Zootaxa. 4392(1):28–40.
- Yang HJ, Kim CH. 2010. Zoeal stages of Lysmata vittata (Decapoda: Caridea: Hippolytidae) reared in the laboratory. Korean J Syst Zool. 26(3):261–278.