Abstract
Barklice in the genus Lepinotus (Psocoptera: Trogiidae) are small, soft-bodied stored-product pests that are difficult to control. We sequenced and annotated the mitochondrial (mt) genome of Lepinotus sp. The mt genome of Lepinotus sp. is 16,299 bp in size with 74.4% A + T content. The gene order was highly conserved in some of the Trogimorpha barklice. Two types of tandem repeat units were identified in CR of Lepinotus sp. The phylogenetic analysis showed that Trogiidae species was the sister group to Lepidopsocidae barklice, and the suborder Troctomorpha was polyphyletic.
Psocoptera (booklice and barklice, also named psocids) contains more than 5000 described species, including three suborders (Trogiomorpha, Psocomorpha and Troctomorpha) (Liu et al. Citation2017). The barklice from genus Lepinotus Heyden, 1850 (Psocoptera: Trogiidae) are stored-product pests worldwide (Arif et al. Citation2012). Lepinotus has more than 12 described species (Liang et al. Citation2012). To date, 13 complete or nearly complete mitochondrial (mt) genomes of barklice species have been reported (Shao et al. Citation2003; Li et al. Citation2013; Liu et al. Citation2017; Yoshizawa et al. Citation2018), and only one incomplete mt genome has been sequenced for Lepinotus species, i.e. Lepinotus reticulatus (Feng et al. Citation2018). To get further information about mt genomes of barklice species, we sequenced and analyzed the complete mt genome of a barklouse, Lepinotus sp.
Lepinotus sp. was collected from Chizhou city, Anhui Province, China in 2014 (30°66′N, 117°49′E). Voucher specimens were deposited in the Entomological Museum (Accession Number: SWU Ps-02-01-01) of the college of Plant Protection, Southwest University. Total genomic DNA was extracted using TIANamp Genomic DNA Kit (Tiangen Biotech, Beijing, China) and stored at −20 °C until future use. Fragments of cox1, cytb, nad5 and 16S were amplified by PCR using conserved insect primers (Simon et al. Citation2006), and nad2 was derived from transcriptome data of this barklouse. Five overlapping fragments (cox1-nad5, nad5-cytb, cytb-16S, 16S-nad2 and nad2-cox1) were amplified by long PCR using the specific primers, which were designed from above-mentioned gene sequences. These fragments were assembled into contigs with SeqMan (DNAStar) and proofed manually. All products were sequenced by the Beijing Genomics Institute (BGI) at Chongqing, China.
The complete mt genome of Lepinotus sp. is a closed-circular molecule of 16,299 bp in size (GenBank accession number: MW735944), containing 13 protein-coding genes (PCGs), 22 tRNA genes, 2 rRNA genes and a control region (CR). Lepinotus sp. has the same gene arrangements and coding strand as in some published Trogimorpha barklice species, e.g. lepidopsocid sp., L. reticulatus and Dorypteryx domestica (Shao et al. Citation2003; Feng et al. Citation2018; Yoshizawa et al. Citation2018). The A + T content of complete mt genome of Lepinotus sp. is 74.4% (A = 38.8%, T = 35.9%, C = 17.9 and G = 7.4%), which is similar to other reported psocids (ranging from 68.6% to 79.8%). All PCGs of Lepinotus sp. initiated with ATD (six ATT, five ATG and two ATA) as the start codons, and mostly terminated with the stop codon TAA/TAG, except for nad5 and cytb (end with the incomplete stop codon T-). The CR (1580 bp) of Lepinotus sp. is located between rrnS and trnQ, with an A + T content of 80.0%. Two types of tandem repeat units (TR1 and TR2) were identified in CR of Lepinotus sp., and the repeat units of TR1 and TR2 were 122 bp (seven times) and 21 bp (six times) in length, respectively. Interestingly, most of the barklice possess tandem repeats in their control region, and these repeat units are highly similar among the species from the same genus, e.g. Lepinotus sp. and L. reticulatus.
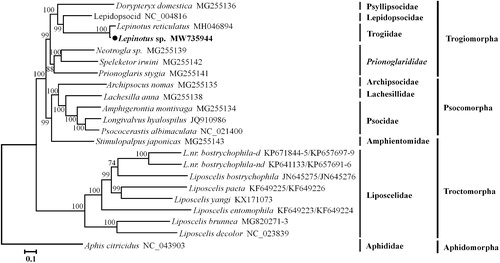
The phylogenetic analysis was performed based on the nucleotide sequences of 11 PCGs (except nad6 and nad4L) and two rRNA genes (in a total length of 11,381 bp) from 20 psocids species. The mt genome sequence of Aphis citricidus was used as an outgroup (Wei et al. Citation2019). A GTR + I + G substitution model for the concatenated dataset was used as the best-fit model, which was determined by jModelTest 2.1.4 (Posada Citation2008). The phylogenetic tree was constructed using the maximum-likelihood (ML) method, which was performed using IQ-TREE web server with 1000 bootstrap replicates (Trifinopoulos et al. Citation2016). The ML tree () consistently supported those of previous mitochondrial phylogenetic studies (Feng et al. Citation2018; Yoshizawa et al. Citation2018). The ML tree showed that each family was recovered as a monophyletic group, and booklice (Liposcelidae) and barklice formed well-supported mono clades, respectively. The suborder Troctomorpha was polyphyletic because the family Amphientomidae was positioned in the suborder Psocomorpha.
Figure 1. Phylogenetic relationships of psocids species inferred from nucleotide sequences of 11 PCGs and two rRNA genes of mitochondrial genomes based on the maximum-likelihood (ML) analysis. Numbers on branches are bootstrap support values. The accession numbers of each species are shown after the scientific name.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in GenBank of NCBI (https://www.ncbi.nlm.nih.gov) with an accession number of MW735944.
Additional information
Funding
References
- Arif M, Ochoa-Corona FM, Opit GP, Li ZH, Kučerová Z, Stejskal V, Yang QQ. 2012. PCR and isothermal-based molecular identification of the stored-product psocid pest Lepinotus reticulatus (Psocoptera: Trogiidae). J Stored Prod Res. 49:184–188.
- Feng S, Stejskal V, Wang Y, Li Z. 2018. The mitochondrial genomes of the barklice, Lepinotus reticulatus and Dorypteryx domestica (Psocodea: Trogiomorpha): insight into phylogeny of the order Psocodea. Int J Biol Macromol. 116:247–254.
- Li H, Shao R, Song F, Zhou X, Yang Q, Li Z, Cai W. 2013. Mitochondrial genomes of two barklice, Psococerastis albimaculata and Longivalvus hyalospilus (Psocoptera: Psocomorpha): contrasting rates in mitochondrial gene rearrangement between major lineages of Psocodea. PLOS One. 8(4):e61685.
- Liang F, Liu X, Zhao J, He Y. 2012. A newly recorded species of the psocid genus Lepinotus Heyden (Psocoptera: Trogiomorpha: Trogiidae) from China. Entomotaxonomia. 34(4):606–608.
- Liu X, Li H, Cai Y, Song F, Wilson JJ, Cai W. 2017. Conserved gene arrangement in the mitochondrial genomes of barklouse families Stenopsocidae and Psocidae. Front Agr Sci Eng. 4(3):358–365.
- Posada D. 2008. jModelTest: phylogenetic model averaging. Mol Biol E. 25(7):1253–1256.
- Shao RF, Dowton M, Murrell A, Barker SC. 2003. Rates of gene rearrangement and nucleotide substitution are correlated in the mitochondrial genomes of insects. Mol Biol E. 20(10):1612–1619.
- Simon C, Buckley TR, Frati F, Stewart JB, Beckenbach AT. 2006. Incorporating molecular evolution into phylogenetic analysis, and a new compilation of conserved polymerase chain reaction primers for animal mitochondrial DNA. Annu Rev Ecol Evol Syst. 37(1):545–579.
- Trifinopoulos J, Nguyen LT, Haeseler A, Minh BQ. 2016. W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 44(W1):W232–W235.
- Wei DD, Lang N, Tao Y, He W, Tu YQ, Miao ZQ, Yang L, Wang JJ. 2019. The mitochondrial genome of the brown citrus aphid Aphis (Toxoptera) citricidus: insights into the repeat regions in aphids and phylogenetic implications. Int J Biol Macromol. 136:531–539.
- Yoshizawa K, Johnson KP, Sweet AD, Yao I, Ferreira RL, Cameron SL. 2018. Mitochondrial phylogenomics and genome rearrangements in the barklice (Insecta: Psocodea). Mol Phylogenet E. 119:118–127.
