Abstract
Quercus chungii F.P.Metcalf, a rare oak with endemic to southern China, belongs to the compound trichome base (CTB) lineage in the Cyclobalanopsis section. The complete chloroplast genome of the species was assembled and annotated in this study. The circular genome was 160,731 bp in size, presenting a typical quadripartite structure including one large single-copy region (LSC, 90,140 bp), one small single-copy region (SSC, 18,911 bp), and two copies of inverted repeat regions (IRs, 25,840 bp). It encoded a total of 113 unique genes, including 79 protein-coding genes, 30 tRNA genes, and four rRNA genes. The maximum-likelihood (ML) phylogenetic tree reconstructed by IQ-TREE indicated that Q. chungii was more closely related to Q. myrsinifolia and Q. sichourensis.
Quercus chungii is a rare and precious tree that is distributed in southern China at elevations ranging from 200 to 800 m. The species belongs to compound trichome base (CTB) lineage in the Quercus section Cyclobalanopsis (Deng et al. Citation2018). With the rapid changes in climate and intensification of human activities, the distribution of Q. chungii is rapidly reduced in recent decades. Understanding the spatial genetic pattern and demographic dynamics of the species can provide important guidelines for the protection and utilization of Q. chungii. The genetic diversity of the species was detected by nuclear simple sequence repeats (SSRs) marker in the previous report (Jiang et al. Citation2019); the spatial genetic pattern and demographic dynamics of Q. chungii from chloroplast DNA remain largely unknown. In this study, we sequenced, assembled, and annotated the complete chloroplast genome of Q. chungii, it could provide useful genomic resources for the future studies on demographic dynamics of the species, and the phylogeny of ring-cupped oaks.
The sample was collected from a wild individual in the Jinpen Mountain National Forest Park, Jiangxi Province, China (25°12′36″N, 115°12′36″E, 566 m). The voucher specimens were deposited in the Herbarium of Shanghai Chenshan Botanical Garden (CSH, http://csh.ibiodiversity.net/default.html, Bin-Jie GE, [email protected], under the voucher number DM12068). Total genomic DNA of sample was extracted from silica-dried leaves using DNeasy plant tissue kit (TIANGEN Biotech Co., Ltd., Beijing, China). Whole genome sequencing was conducted with the Illumina Hiseq X Ten platform. A total of 66,910,243 clean reads were produced and 50,000,000 reads were used for the de novo assembly with GetOrganelle v1.7.2beta (Jin et al. Citation2020). Gene annotation was performed by the pipeline PGA (Qu et al. Citation2019).
The complete plastid genome of Q. chungii was a circular molecule of 160,731 bp in length. It had a typical quadripartite structure including one large single-copy (LSC) region (90,140 bp), one small single-copy (SSC) region (18,911 bp), and two copies of inverted repeat (IRs) regions (25,840 bp). The overall GC content was 36.91%, while the corresponding values of the LSC, SSC, and IR regions were 34.76%, 31.11%, and 42.77%, respectively. A total of 130 genes were encoded, of which 113 were unique and 17 were duplicated in the IR regions. Among the unique genes, 79 were protein-coding genes, 30 were tRNA genes, and four were rRNA genes.
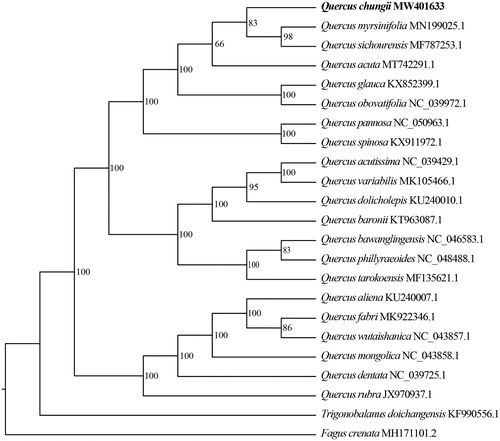
To identify the phylogenetic position of Q. chungii, a maximum-likelihood (ML) tree was reconstructed based on the chloroplast genome sequences of Q. chungii and relatives species. Two species, Trigonobalanus doichangensis and Fagus crenata, were selected as outgroups. The sequences were aligned by MAFFT 7.475 (Rozewicki et al. Citation2019). The ML analyses were performed with IQ-TREE 1.6.12 (Chernomor et al. Citation2016). Node support was assessed by 1000 fast bootstrap replicates. Our result indicated that Q. chungii was more closely related to Q. myrsinifolia and Q. sichourensis with 83% bootstrap support ().
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The complete chloroplast genome sequence of Quercus chungii is deposited in the GenBank database under the accession number MW401633 (https://www.ncbi.nlm.nih.gov/nuccore/MW401633). Raw sequencing reads used in this study were deposited in the public repository BioSample with accession number SAMN18499615 (https://www.ncbi.nlm.nih.gov/biosample/SAMN18499615).
Additional information
Funding
References
- Chernomor O, von Haeseler A, Minh BQ. 2016. Terrace aware data structure for phylogenomic inference from supermatrices. Syst Biol. 65(6):997–1008.
- Deng M, Jiang XL, Hipp AL, Manos PS, Hahn M. 2018. Phylogeny and biogeography of East Asian evergreen oaks (Quercus section Cyclobalanopsis; Fagaceae): insights into the Cenozoic history of evergreen broad-leaved forests in subtropical Asia. Mol Phylogenet Evol. 119:170–181.
- Jiang XL, Xu GB, Deng M. 2019. Spatial genetic patterns and distribution dynamics of the Rare Oak Quercus chungii: implications for biodiversity conservation in Southeast China. Forests. 10(9):821.
- Jin JJ, Yu WB, Yang JB, Song Y, dePamphilis CW, Yi TS, Li DZ. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):241.
- Qu XJ, Moore MJ, Li DZ, Yi TS. 2019. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods. 15(1):50.
- Rozewicki J, Li S, Amada KM, Standley DM, Katoh K. 2019. MAFFT-DASH: integrated protein sequence and structural alignment. Nucleic Acids Res. 47(W1):W5–W10.