Abstract
The genus Lonicera (Caprifoliaceae) is of great economical significance. It has been taxonomically studied frequently in history, while phylogenetic relationships intra the genus are still obscure. Here, we reported the first species complete chloroplast genome sequence in the section Isoxylosteum, Lonicera angustifolia var. myrtillus. It is 156,222 bp in length, comprising a large single-copy (LSC) region of 89,838 bp, a small single-copy (SSC) region of 19,211 bp, and a pair of inverted repeats (IRs) of 23,509 bp. In L. angustifolia var. myrtillus chloroplast genome, a total of 114 functional genes were identified, with an overall GC content of 38.4%. The phylogenetic relationships of Lonicera based on maximum-likelihood (ML) showed that L. angustifolia var. myrtillus is most closely related to L. nervosa in section Isika. Our study contributes to the molecular phylogenetic studies of Lonicera and Caprifoliaceae.
The genus Lonicera (Caprifoliaceae) have significant ornamental, medicinal, and edible value. There are approximately 200 species in Lonicera which are mainly distributed in temperate and subtropical areas in the Northern Hemisphere, with several species extending their range into tropical areas of India, Malaysia, Philippines, and North Africa (Rehder Citation1903, Citation1913; van Steenis Citation1946; Theis et al. Citation2008), some of them are highly endangered, such as L. oblata (Zhu et al. Citation2019; Wu et al. Citation2021). Historically, Lonicera has received extensive taxonomic evaluation and phylogenetic inference, while its phylogenetic relationships among sections, subsections, and species are still obscure (Theis et al. Citation2008; Nakaji et al. Citation2015).
Lonicera angustifolia var. myrtillus (Hook. f. & Thomson) Q. E. Yang, Landrein, Borosova & J. Osborne, a subalpine deciduous shrub with axillary pinkish white or purple-red paired flowers, occurs in shrubby hillside, sparse forests, and stony places along valleys with an altitude of 2400–4000 m. It belongs to the section Isoxylosteum, in which no complete plastome is reported so far. In order to better understand the molecular phylogenetic relationship of Lonicera, we reported and characterized the first complete chloroplast genome of L. angustifolia var. myrtillus using the next-generation sequencing technology. Furthermore, a phylogenomic analysis of this species and its relatives was also presented.
Fresh young leaves of L. angustifolia var. myrtillus were collected from Galongla Mountain, Bomi County, Xizang Autonomous Region (29.7643°N, 95.6975°E). Voucher specimen was preserved in PE (collector and collection number: Bing Liu ([email protected]), Jian-Fei Ye & Hong-Qiang Xiao 3850). Genomic DNA extraction and next-generation sequencing were performed with an Illumina Hiseq platform by Shanghai OE Biotech. Co., Ltd. (http://oebiotech.bioon.com.cn/, Shanghai, China). We used Map function of Geneious R11 (Kearse et al. Citation2012) to select chloroplast reads using published chloroplast genome of L. japonica as reference (Kang et al. Citation2018). Then, these filtered reads were de novo assembled with Geneious R11. Gaps were filled using Fine Tuning function of Geneious R11. The assembled chloroplast sequence was then annotated using the Plann (Huang and Cronk Citation2015). Eventually, annotations were verified by Geneious R11.
The complete chloroplast genome sequence of L. angustifolia var. myrtillus is with 156,222 bp in length as a circle, which has a characteristic quadripartite structure with a large single-copy (LSC) region of 89,838 bp, a small single-copy (SSC) region of 19,211 bp, and a pair of inverted repeats (IRs) of 23,509 bp. In L. angustifolia var. myrtillus chloroplast genome, a total of 114 functional genes were identified, including 80 protein-coding genes (PCGs), 30 tRNA genes, and four rRNA genes. The total sequence GC content of the complete plastome is 38.4%.
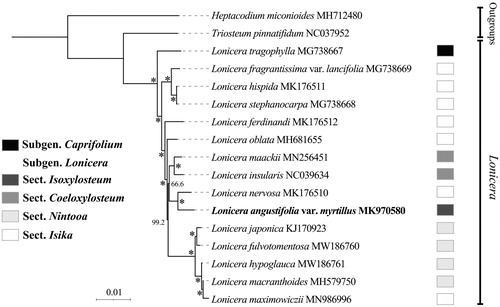
Species phylogeny intra Lonicera and the phylogenetic position of L. angustifolia var. myrtillus were evaluated based on whole plastome data. The complete chloroplast genome sequences of L. angustifolia var. myrtillus and other 16 species from Lonicera, with Heptacodium miconioides and Triosteum pinnatifidum from Caprifoliaceae as outgroup were used for phylogenetic analysis. A total of 19 complete chloroplast genomes were aligned using MAFFT (Katoh et al. Citation2005). The phylogenetic tree was reconstructed and analyzed by the maximum-likelihood method with 1000 bootstrap values replicate at each node based on GTR + G model in RAxML software (Kozlov et al. Citation2019). The final tree was edited using the iTOL version 5.0 online web (https://itol.embl.de/) (Letunic and Bork Citation2016). A robust phylogeny of Lonicera was obtained based on the whole plastome data, and the genus was resolved as a monophyletic clade consisting two well supported clades, subgen. Lonicera and subgen. Caprifolium (). Our result supports the classification of the two subgenera in Lonicera proposed by Rehder (Citation1903, Citation1913), Hara (Citation1983), and Hsu and Wang (Citation1988) and coincides with previous molecular phylogenetic studies (Theis et al. Citation2008; Smith Citation2009; Nakaji et al. Citation2015; Zhu et al. Citation2019). However, all the four sections (sect. Coeloxylosteum, sect. Isika, sect. Isoxylosteum, and sect. Nintooa) classified in the subgen. Lonicera proposed by Rehder (Citation1903, Citation1913) and Hsu and Wang (Citation1988) were not supported as monophyly, all three of these sections are nested within the section Isika (Theis et al. Citation2008; Nakaji et al. Citation2015; Zhu et al. Citation2019), indicating that phylogenetic relationship intra subgen. Lonicera is complicated and not exactly consistent with traditional morphology. More species of this subgenus are needed to further investigate its phylogeny. As the first species whose whole plastome was reported in sect. Isoxylosteum, L. angustifolia var. myrtillus was indicated closely related to L. nervosa in a polyphyletic group, sect. Isika, which is consistent with previous study based on nuclear and chloroplast DNA sequences by Theis et al. (Citation2008). Our results provide valuable data and shed light on the phylogenomic study of Lonicera.
Figure 1. A phylogenetic tree of Lonicera L. inferred from the maximum-likelihood method based on the complete chloroplast genome data. *The clade support value of 100 that was generated from the maximum likelihood method. The taxonomic groups are following Rehder (Citation1903, Citation1913), Hara (Citation1983), and Hsu and Wang (Citation1988).
Disclosure statement
No potential conflict interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under the accession MK970580. Raw Illumina data were deposited in the NCBI Sequence Read Archive (SRA: SUB8882578, BioProject: PRJNA691677, and Bio-Sample: SAMN17302025).
Additional information
Funding
References
- Hara H. 1983. A revision of Caprifoliaceae of Japan with reference to allied plants in other districts and the Adoxaceae. Ginkgoana: contributions to the Flora of Asia and the Pacific Region. Tokyo: Academia Scientific Book Inc.; p. 336.
- Hsu PS, Wang HJ. 1988. Lonicera. In: Hsu PS, editor. Flora Reipublicae Popularis Sinicae. Vol. 72. Beijing: Science Press; p. 143–259.
- Huang DI, Cronk QC. 2015. Plann: a command-line application for annotating plastome sequences. Appl Plant Sci. 3(8):1500026.
- Kang S, Park H, Koo HJ, Park JY, Lee DY, Kang KB, Han SI, Sung SH, Yang T. 2018. The complete chloroplast genome sequence of Korean Lonicera japonica and intra-species diversity. Mitochondrial DNA B Resour. 3(2):941–942.
- Katoh K, Kuma KI, Toh H, Miyata T. 2005. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 33(2):511–518.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Kozlov AM, Darriba D, Flouri T, Morel B, Stamatakis A. 2019. RAxML-NG: a fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics. 35(21):4453–4455.
- Letunic I, Bork P. 2016. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 44(W1):W242–W245.
- Nakaji M, Tanaka N, Sugawara T. 2015. A molecular phylogenetic study of Lonicera L. (Caprifoliaceae) in Japan based on chloroplast DNA sequences. Acta Phytotaxon Geobot. 66(3):137–151.
- Rehder A. 1903. Synopsis of the genus Lonicera. Missouri Bot Gard Ann Rep. 1903:27–232.
- Rehder A. 1913. Caprifoliaceae. In: Sargent CS, editor. Plantae Wilsonianae: an enumeration of the woody plants collected in western China for the Arnold Arboretum of Harvard University during the years 1907, 1908, and 1910. Cambridge: Harvard University Press; p. 106–144.
- Smith SA. 2009. Taking into account phylogenetic and divergence‐time uncertainty in a parametric biogeographical analysis of the Northern Hemisphere plant clade Caprifolieae. J Biogeogr. 36(12):2324–2337.
- Theis N, Donoghue MJ, Li JH. 2008. Phylogenetics of the Caprifolieae and Lonicera (Dipsacales) based on nuclear and chloroplast DNA sequences. Syst Bot. 33(4):776–783.
- van Steenis CGGJ. 1946. Preliminary revision of the genus Lonicera in Malaysia. J Arnold Arboretum. 27:442–452.
- Wu YM, Shen XL, Tong L, Lei FW, Mu XY, Zhang ZX. 2021. Impact of past and future climate change on the potential distribution of an Endangered montane shrub Lonicera oblata and its conservation implications. Forests. 12(2):125.
- Zhu YX, Wu YM, Shen XL, Tong L, Xia XF, Mu XY, Zhang ZX. 2019. The complete chloroplast genome of Lonicera oblata, a critically endangered species endemic to North China. Mitochondrial DNA B Resour. 4(2):2337–2338.
