Abstract
Zanthoxylum bungeanum var. pubescensis is a variety species of Z. bungeanum Maxim. that is characterized with small leaf plants and possess high resistant to cold stress among the genus Zanthoxylum. In the current study, the complete chloroplast genome sequences of two different leaf shapes were assembled. One type of leaflets is thin paper, and the color on both sides is obviously different after drying, and the fruit stalk is slender and elongated; the other type of leaflets are the thick paper, the leaf surface and fruit stalk are glabrous, the fruit stalk is thicker, and the lateral veins are recessed on the leaf surface and appear to be cracked. Their circular DNA lengths were all 158,565 bp. Both genomes were made up of a large single-copy (LSC), a small single-copy (SSC), and two inverted repeat (IRs) regions. Each genome encoded 132 genes, including 87 protein-coding, 37 tRNA, and 8 rRNA genes. Phylogenetic analysis indicated that both species were closely related to the congeneric Z. bungeanum.
Zanthoxylum bungeanum var. pubescens Huang is distributed in Qinghai, Gansu, southern Shaanxi, western Sichuan, and northwest Sichuan in China, and it is wild in the valley, forest edge and gully edge, Zanthoxylum bungeanum var. pubescens belongs to the Rutaceae family and is a kind of small leaf plant (Editing Committee of Chinese Flora Citation1993). It is one of cold-resistant species in the genus Zanthoxylum. We found an extremely small Z. bungeanum var. pubescens population, which with different leaf shapes in the field surveys. Up to date, the information of the complete chloroplast genome of Z. bungeanum var. pubescens has not been reported. Therefore, it is necessary to develop genomic resources for Z. bungeanum var. pubescens to provide more basic information, and to promote its systematic research and conservation.
The leaves of Z. bungeanum var. pubescens were collected from two wild plants in Aba Tibetan and Qiang Autonomous Prefecture, Sichuan, China (31.32°N, 103.40°E). The voucher specimen (MYHJ1 and MYHJ2) were deposited at the Herbarium College of Landscape Architecture and Life Science, Chongqing University of Arts and Sciences, Chongqing, China. The total DNA was extracted from fresh leaves using a modified CTAB method (Doyle and Doyle Citation1987). The whole chloroplast genome sequencing on the BGISEQ-500 platform was performed by Bio&Data Biotechnology Co. Ltd. (Hefei, China). We have obtained 43,063,984 raw reads (MYHJ1) and 41,217,366 raw reads (MYHJ2), and the percentage of raw reads gathered as clean reads were 98.5% and 98.0%, respectively. The filtered reads were assembled using the program SPAdes assembler 3.9.0 (Bankevich et al. Citation2012). The chloroplast genome was annotated using CpGAVAS (Liu et al. Citation2012) and GeSeq software (Tillich et al. Citation2017). Annotation errors were corrected manually. The raw reads were deposited in the NCBI Sequence Read Archive (SRA: SRX9591242 and SRX9591243) and the final annotated chloroplast genome sequence has been submitted to the NCBI database (accession numbers: MW206783 and MW206784).
The size of two circular chloroplast genome sequences was 158,565 bp, a large single-copy (LSC) region of 85,746 bp and a small single-copy (SSC) region of 17,597 bp separated by a pair of identical inverted repeat regions (IRa and IRb) of 27,611 bp each. Both genomes encoded 132 genes, including 87 protein-coding, 37 tRNA, and 8 rRNA genes. The GC content of the two whole genomes, LSC, SSC, and IRs regions were 38.5, 36.8, 33.5, and 42.5%, respectively. GC content of IRs region was the highest. Among the genes, 8 protein-coding, 7 tRNA, and 4 rRNA genes were found duplicated in IR regions. Nineteen genes contain two exons, while four genes (clpP, ycf3, and two rps12) have three exons. A base mutation between these two samples occurred in the intergenic region of trnE-UUC and trnT-GGU of tRNA. It also exhibited compared with Z. bungeanum var. pubescens, 190 mutation loci including SNP and indel were found in Z. bungeanum (NC_031386), of which the largest indel is located inside the rpl22 gene. Additionally, at the junction of LSC and IRB, 153 bp deletion was observed in Z. bungeanum compared with that of Z. bungeanum var. pubescens.
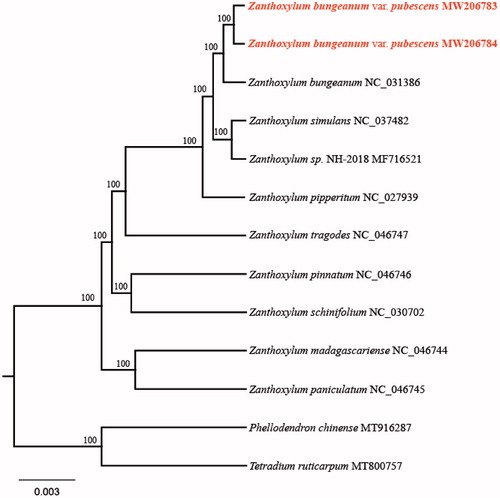
To confirm the phylogenetic relationship of both species with other members of the genus Zanthoxylum, we used complete chloroplast genome sequences of 12 species, and constructed a phylogenetic tree (). These sequences were aligned with MAFFT v7.407 (Katoh and Standley Citation2013). The maximum likelihood (ML) tree was performed with RAxML version 8 program (Alexandros Citation2014) using 1000 bootstrap. The result showed that both Z. bungeanum var. pubescens species were closely related to Z. bungeanum with bootstrap support values of 100%.
Acknowledgments
We thank Yinming Wu and Wei Tang (Sichuan Academy of Botanical Engineering, China) for their help in sample collections.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/), with accession number [MW206783; MW206784]. The associated SRA number are SRX9591242 and SRX9591243.
Additional information
Funding
References
- Alexandros S. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19(1):11–15.
- Editing Committee of Chinese Flora. 1993. Flora of China. Beijing (China): Science Press; p. 46.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Liu C, Shi L, Zhu Y, Chen H, Zhang J, Lin X, Guan X. 2012. CpGAVAS, an integrated web server for the annotation, visualization, analysis, and GenBank submission of completely sequenced chloroplast genome sequences. BMC Genom. 13(1):715.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq - versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.