Abstract
Orixa japonica Thunb. is an important medicinal plant belonging to the family Rutaceae. In this study, we determined the the complete chloroplast (cp) genome of O. japonica, which was 158,525 bp in length containing one large single copy region (85,965 bp), one small single copy region (18,552 bp), and a pair of inverted repeat regions (27,004 bp each). A total of 134 genes were annotated in the cp genome, including 88 protein coding genes, 37 tRNA genes, eight rRNA genes, and one pseudo gene ycf1. According to the phylogenetic analysis, O. japonica clustered together with Casimiroa edulis with high bootstrap value, indicating a close genetic relationship with subfamily Amyridoideae.
Orixa japonica Thunb. (O. japonica) is a shrub or tree belonging to the family Rutaceae that has noteworthy medicinal value and is worth doing profound researches. It is widely distributed in southern China (Thunb Citation1997). The roots and stems of it are used as a kind of herbal medicine in Chinese folk. According to records, it has certain toxicity and can clear heat and dampness, relieve cough, analgesia, and emetic. It is used to treat stomachache, rheumatic arthralgia, malaria, etc. The extracts or constituents of it possess pharmacological and biological activities. Previous chemical studies have isolated and identified several quinoline alkaloids, terpenoids, and coumarins from it and pyrrolidine alkaloid isolated from root bark of the O. japonica has potential nematicidal and larvicidal activity that can be developed as natural nematicides and larvicides in the future (Liu et al. Citation2016). However, the genomic information of O. japonica has not been established sufficiently, the complete chloroplast (cp) genome of O. japonica has not been sequenced, which limits the in-depth study of it. The complete cp genome contains a lot of molecular information that is a useful tool for phylogenetic analysis and further development on species identification as well as conservation strategies. Therefore, it is worthwhile to sequence the complete cp genome of O. japonica and analyze characters of it to pave the way for its further genetics research.
In this study, the complete cp genome of O. japonica has been determined and its phylogenetic relationship within family Rutaceae also has been analyzed through the complete cp genome. Total genomic DNA was extracted from fresh leaves of O. japonica specimen by modified CTAB method (Doyle and Doyle Citation1987). The specimen was collected from Fuyang District, Hangzhou City, Zhejiang Province (30°04′59.74″N, 119°53′10.14″E) and deposited in the Medicinal Herbarium Center of Zhejiang Chinese Medical University, Hangzhou, Zhejiang, China (voucher identifying number CCSZK-200816). Illumina Hiseq Platform (Illumina, San Diego, CA) was used to sequence the total genomic DNA and metaSPAdes (Nurk et al. Citation2017) was used to assemble the complete cp genome of O. japonica with the complete cp genome of Phellodendron amurense (GenBank accession number: NC_035551) as reference (Dong et al. Citation2020; Gao et al. Citation2020). The complete cp genome of O. japonica was annotated using Geseq (Tillich et al. Citation2017) and then was manually confirmed by BLAST. The final annotated cp genome of O. japonica was submitted to GenBank with the accession number MW574915.
The complete cp genome of O. japonica in total is 158,525 bp in length and consists of a large single copy region (LSC; 85,965 bp), a small single copy region (SSC; 18,552 bp), and a pair of inverted repeat regions (IRs; 27,004 bp each). There are a total of 134 genes annotated in the cp genome, including 88 protein coding genes, 37 tRNA genes, eight rRNA genes, and one pseudo gene ycf1. In addition, the overall GC content of the complete cp genome is 38.21%, and the corresponding values for LSC, SSC, and IR regions are 36.49%, 32.68%, and 42.86%, respectively. There are 21 duplicated genes in the IR region.
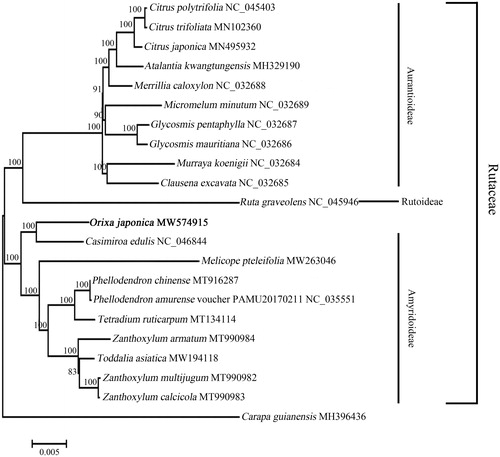
In order to perform the phylogenetic analysis, a maximum-likelihood (ML) tree was constructed using the complete cp genome sequence of O. japonica and 20 other species from family Rutaceae, Carapa guianensis was selected as the outgroup. These complete cp genome sequences were downloaded from NCBI and first aligned using MAFFT v7.037b (Katoh and Standley Citation2013), then the ML tree was constructed by MEGA 7 (Kumar et al. Citation2016) with 100 bootstrap replications. According to the ML tree (), O. japonica clustered together with Casimiroa edulis with high bootstrap value, indicating that they have close relationship. In Flora Reipublicae Popularis Sinicae, genera Ruta, Orixa, Melicope, Tetradium, and Zanthoxylum belong to the subfamily Rutoideae (Engl Citation1997a, Citation1997b), genera Casimiroa, Phellodendron, and Toddalia belong to the subfamily Toddalioideae (Engl Citation1997a, Citation1997b). However, in the most recent classification of Rutaceae, Amyridoideae is the most diverse subfamily, genera Casimiroa, Melicope, Phellodendron, Tetradium, Toddalia, and Zanthoxylum belong to the subfamily Amyridoideae (Morton and Telmer Citation2014; Sun et al. Citation2021), but which subfamily genus Orixa belongs to is still not clear. According to our result, O. japonica formed a clade with the plants from subfamily Amyridoideae, suggesting that O. japonica might has close genetic relationship with subfamily Amyridoideae. These findings provide fundamental valuable molecular information for O. japonica and laid a foundation for its identification, as well as further research on genetics of family Rutaceae in the future.
Figure 1. Phylogenetic relationships of Orixa japonica and 20 other species from family Rutaceae. The maximum-likelihood (ML) tree was constructed by MEGA 7 based on Kimura 2-parameter model using the complete chloroplast genomes. Carapa guianensis, which was from family Meliaceae, was selected as the outgroup. Numbers on the nodes represent bootstrap values from 100 replicates. The GenBank accession numbers were listed following the species name.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under the accession no. MW574915. The associated BioProject, SRA, and BioSample numbers of Orixa japonica are PRJNA702044, SRR13717521, and SAMN17922835, respectively.
Additional information
Funding
References
- Dong S, Gao C, Wang Q, Ge Y, Cheng R. 2020. Characterization of the complete chloroplast genome of Macleaya cordata and its phylogenomic position within the subfamily Papaveroideae. Mitochondrial DNA B Resour. 5(2):1714–1715.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Engl. 1997a. Subfam. Rutoideae Engl. Flora Reipublicae Popularis Sinicae. Vol. 43. Beijing: Science Press; p. 8.
- Engl. 1997b. Subfam. Toddalioideae Engl. Flora Reipublicae Popularis Sinicae. Vol. 43. Beijing: Science Press; p. 96.
- Gao C, Wang Q, Ying Z, Ge Y, Cheng R. 2020. Molecular structure and phylogenetic analysis of complete chloroplast genomes of medicinal species Paeonia lactiflora from Zhejiang Province. Mitochondrial DNA B Resour. 5(1):1077–1078.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kumar S, Stecher G, Tamura K. 2016. Mega 7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Liu X, Lai D, Liu Q, Zhou L, Liu Q, Liu Z. 2016. Bioactivities of a new pyrrolidine alkaloid from the root barks of Orixa japonica. Molecules. 21(12):1665.
- Morton CM, Telmer C. 2014. New subfamily classification for the Rutaceae. Ann Missouri Bot Gard. 99(4):620–641.
- Nurk S, Meleshko D, Korobeynikov A, Pevzner PA. 2017. metaSPAdes: a new versatile metagenomic assembler. Genome Res. 27(5):824–834.
- Sun K, Liu QY, Wang A, Gao YW, Zhao LC, Guan WB. 2021. Comparative analysis and phylogenetic implications of plastomes of five genera in subfamily Amyridoideae (Rutaceae). Forests. 12(3):277.
- Thunb. 1997. Orixa japonica Thunb. Flora Reipublicae Popularis Sinicae. Vol. 43. Beijing: Science Press; p. 54.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq- versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
