Abstract
Alternanthera philoxeroides (Mart.) Griseb. (Alternanthera philoxeroides) is an important herbage species, which could provide high-quality feed for livestock and poultry breeding. This paper is the first to report the A. philoxeroides’s chloroplast genomes, which were detected by de novo sequencing. The results showed that the length of A. philoxeroides’ chloroplast genome sequence was 152,255 bp, including a large single-copy (LSC) region (84,670 bp), a small single-copy (SSC) region (17,343 bp), and two inverted repeat (IR) regions (25,121 bp). Alternanthera philoxeroides’ chloroplast genome encoded 132 genes including 8 rRNA, 38 tRNA, and 86 protein-coding genes. After phylogenetic and cluster analysis, A. philoxeroides was closest to Amaranthaceae, and the relationship between Amaranthus and Achyranthes was closest.
Alternanthera philoxeroides (Mart.) Griseb. (Alternanthera philoxeroides) is an Amaranthaceae family originating in South America, and it is all over the world now (Telesnicki et al. Citation2011; Jin et al. Citation2021). Alternanthera philoxeroides has enlisted the Preliminary list of alien invasive species by the Ministry of Ecology and Environment of the People’s Republic of China in 2002. Although Alternanthera philoxeroides are a harmful species that invasive to China, it is used as herbage for animals’ breeding at present (Pan et al. Citation2007; Tan and Zhang Citation2007; Wang and Huang Citation2011). Alternanthera philoxeroides is very rich in wild resources spreading over the marshes, rivers, and lakes in China (Li et al. Citation2012; Yan et al. Citation2020). Thus it is a very promising industry to make rational use of the resources of Alternanthera philoxeroides to provide high-quality feed for livestock and poultry breeding. However, there is little research on the resource utilization of Alternanthera philoxeroides, especially the comprehensive utilization and development of its genetic resources and species resources. Since the chloroplast genome has conserved structure and orthologous comparing with the nuclear genome, it plays a pivotal role in Alternanthera philoxeroides’ genetics and evolution (Cui et al. Citation2006; Zuo et al. Citation2019; Yinran et al. Citation2020). Nevertheless, there is no relevant chloroplast genome research about the Alternanthera philoxeroides leading the process of molecular genetics research was been prevented. Therefore, the complete chloroplast genomes of Alternanthera philoxeroides was been detected by de novo sequencing.
The fresh leaves of Alternanthera philoxeroides was been collected from Pihe National Wetland Park, Lu’an city, Anhui province, P. R. China (N:31.76°, E:116.49°), and the fresh leaves of Alternanthera philoxeroides was deposited at West Anhui University (Ping Jiang, [email protected]) under the voucher number TCVM202008280035. The Plant DNA extraction kit was used to extract the total DNA of Alternanthera philoxeroides. After the DNA quality meets the sequencing requirements detected by the method of micro-volume spectrophotometer and 1% agarose electrophoresis, the DNA was escorted to Beijing Zhongxingbomai Technology for sequencing using Illumina NovaSep platform.
The Raw data were filtered and Get Organelle pipeline (Jin et al. Citation2020) was run to obtain high-quality data. The contig sequence was been assembled by SPAdes de novo software (Luo et al. Citation2012; Yinran et al. Citation2020), and the relative position of the genome in the contig sequences was acquired by the BLAT (Kent Citation2002) referencing the NCBI database (NC 024157.1, NC 011942.1, NC 009618.1, NC 000932.1, KX 352464.1). The Bandage tool (Wick et al. Citation2015) and the Geseq program (Tillich et al. Citation2017) were run to obtain the full-length frame diagram and annotate the chloroplast genome, respectively. The OGDRAW software (Lohse et al. Citation2013) was run to draw the physical map (GenBank accession number: MW285080).
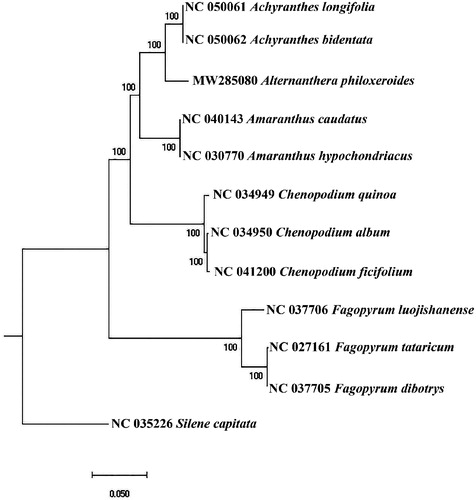
Similar to other higher plants, the Alternanthera philoxeroides has a typical four-segment structure, including a large single-copy (LSC) region (84,670 bp), a small single-copy (SSC) region (17,343 bp), and two inverted repeat (IR) regions (25,121 bp). And the plastome sequence of Alternanthera philoxeroides’ chloroplast genome was 152,255 bp. Its GC content was 36.40% and contained 132 genes including 8 rRNA, 38 tRNA, and 86 protein-coding genes. To determine the phylogenetic status of Alternanthera philoxeroides in Amaranthaceae plants, we used two Amaranthus, two Achyranthes, three Chenopodium, and three Fagopyrum plants and 1 outgroup (Silene) in NCBI for phylogenetic analysis. The GTRCATX model from raxmlGUI version 1.5 b (Silvestro and Michalak Citation2012) performed 1000 bootstrap replicates to calculate the evolutionary relationship and drawing the phylogenetic tree. The cluster analysis results showed that Alternanthera philoxeroides was closest to Amaranthaceae, and the relationship between Amaranthus and Achyranthes was closest ().
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study have send it up to BankIt (2403319) of National Center for Biotechnology Information, and provided GenBank accession number (MW285080). The associated BioProject and Bio-Sample numbers are PRJNA683712 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA683712) and SRX9689754 (https://www.ncbi.nlm.nih.gov/sra/SRX9689754]), respectively.
Additional information
Funding
References
- Cui LY, Veeraraghavan N, Richter A, Wall K, Jansen RK, Leebens-Mack J, Makalowska I, dePapmphilis CW. 2006. ChloroplastDB: the Chloroplast Genome Database. Nucleic Acids Res. 34:692–696.
- Huang YR, Huang XX, Feng SX, Yan SF, Li YT, Liu YC. 2020. The complete chloroplast genome sequence of Ulmus elongata (Ulmaceae). Mitochondrial DNA Part B. 5:792–793.
- Jin JJ, Yu WB, Yang JB, Song Y, dePamphilis CW, Yi TS, Li DZ. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21:241.
- Jin JS, Zhao MT, Zhang H, Liu YR, Wan FH, Zhou ZS, Guo JY. 2021. Impact of summer temperatures on Agasicles hygrophila, a key biocontrol agent of the invasive weed Alternanthera philoxeroides in Hunan province, China. Entomologia Generalis. 41:59–70.
- Kent WJ. 2002. BLAT-the BLAST-like alignment tool. Genome Res. 12:656–664.
- Li RZ, Li F, Zhou AJ. 2012. Sediment and pore water nutrient characteristics in growing zones of Alternanthera philoxeroides in the Shiwuli River, Chaohu Lake. Env Sci. 33:3014–3023. in Chinese.
- Lohse M, Drechsel O, Kahlau S, Bock R. 2013. OrganellarGenomeDRAW-a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 41:W575–W581.
- Luo RB, Liu BH, Xie YL, Li ZY, Huang WH, Yuan JY, He GZ, Chen YX, Pan Q, Liu YJ, Tang JB, Wu GX, Zhang H, Shi YJ, Liu Y, Yu C, Wang B, Lu Y, Han CL, Cheung DW, Yiu SM, Peng SL, Zhu XQ, Liu GM, Liao XK, Li YR, Yang HM, Wang J, Lam TW, Wang J. 2012. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience. 1:18.
- Pan XY, Geng YP, Sosa A, Zhang WJ, Li B, Chen JK. 2007. Invasive Alternanthera philoxeroides: biology, ecology and management. J Syst Evol. 45:884–900. in Chinese.
- Silvestro D, Michalak I. 2012. raxmlGUI: a graphical front-end for RAxML. Org Divers Evol. 12:335–337.
- Tan P, Zhang XJ. 2007. The chemical composition and application of Alternanthera philoxeroides. J Public Health Prev Med. 18:50–52. in Chinese.
- Telesnicki MC, Sosa AJ, Greizerstein E, Julien MH. 2011. Cytogenetic effect of Alternanthera philoxeroides (alligator weed) on Agasicles hygrophila (Coleoptera: Chrysomelidae) in its native range. Biol Control. 57:138–142.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq-versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45:6–11.
- Wang GY, Huang GH. 2011. Feeding lactating dairy cows with Alternanthera philoxeroides. Shanghai J Animal Husb Vet Med. 4:32–33. in Chinese.
- Wick RR, Schultz MB, Zobel J, Holt KE. 2015. Bandage: interactive visualization of de novo genome assemblies. Bioinformatics. 31:3350–3352.
- Yan HY, Feng L, Zhao YF, Feng L, Wu D, Zhu CP. 2020. Prediction of the spatial distribution of Alternanthera philoxeroides in China based on ArcGIS and MaxEnt. Global Ecol Conserv. 21:e00856.
- Zuo RH, Sun CB, Chen CW, Jiang P. 2019. Complete chloroplast genome of Rhus chinensis by de novo sequencing. Mitochondrial DNA Part B. 4:1672–1673.