Abstract
The chloroplast genome of Abeliophyllum distichum f. lilacinum Nakai, classified to a monotypic in this genus, and an endemic species in Korea, was sequenced to understand the genetic differences among intraspecies and cultivars of A. distichum. The chloroplast genome length is 156,015 bp (GC ratio is 37.8%) and has a typical quadripartite structure: 86,779 bp large single copy (35.8%) and 17,828 bp small single copy (31.9%) regions separated by two 25,704 bp inverted repeat (43.2%) regions. The genome encodes for 133 genes (88 protein-coding genes, eight rRNAs, and 37 tRNAs). Six to 99 SNPs and seven to 18 INDEL regions (19 bp to 72 bp) were identified against available chloroplast genomes of A. distichum. Phylogenetic trees show that A. distichum f. lilacinum is clustered with the Dae Ryun cultivar which has a larger fruit body. Our analyses suggest additional research, such as Genotyping-By-Sequencing, for understanding relationship between morphology and genotype of A. distichum.
Abeliophyllum distichum Nakai, a monotypic species in Abeliophyllum (Nakai Citation1919; Melchior Citation1964), is an endemic species in Korea (Park et al. Citation2020). It presents diverse morphological features such as leaves and flowers, resulting in the four forma: i) A. distichum f. lilacinum, pink-colored flower, ii) A. distichum f. edburneum, ivory-colored flower, iii) A. distichum f. viridicalycinum, light-green-colored flower, and iv) A. distichum f. rotundicarpum, round-shaped fruit (Lee Citation1976). Due to these morphological variations, one cultivar was developed and at least two more cultivars are being developed for horticultural purposes (Min et al. Citation2019; Park et al. Citation2019; Park et al. Citation2019). Moreover, A. distichum contains useful phytocompounds including acteoside (Jang et al. Citation2020; Park et al. Citation2020) and hirsutrin (Oh et al. Citation2003). Here, we sequenced and reported the chloroplast genome of A. distichum f. lilacinum to better understand its intraspecific genetic diversity.
Fresh leaves material was collected in GoesanBunjae-Nongwon in Goesan-gun, Chungbuk Province, Republic of Korea (36.797369 N, 127.782997E). Total genomic DNA was extracted from the collected leaves material using the DNeasy Plant Mini Kit (QIAGEN, Hilden, Germany). The voucher was deposited in the InfoBoss Cyber Herbarium (IN; http://herbarium.infoboss.co.kr/; Voucher number: IB-01092; Contact: Suhyeon Park; [email protected]). Genome sequencing was conducted using NovaSeq6000 at Macrogen Inc., Korea, and de novo assembly and sequence confirmation were conducted using Velvet v1.2.10 (Zerbino and Birney Citation2008), GapCloser v1.12 (Zhao et al. Citation2011), BWA v0.7.17 (Li Citation2013), and SAMtools v1.9 (Li et al. Citation2009) under the environment of Genome Information System (GeIS; http://geis.infoboss.co.kr/). Geneious Prime® v2020.2.4 (Biomatters Ltd., Auckland, New Zealand) was used for chloroplast genome annotation based on the A. distichum chloroplast genome (MN116559; Park, Min, et al. Citation2019).
The chloroplast genome of A. distichum f. lilacinum (GenBank accession is MW426545) is 156,015 bp (GC ratio is 37.8%) and has four subregions: 86,779 bp of large single copy (GC ratio is 35.8%) and 17,828 bp of small single copy (GC ratio is 31.9%) regions are separated by two 25,704 bp of inverted repeat (IR; GC ratio is 43.2%). The chloroplast genome contains 133 genes consisting of 88 protein-coding genes, eight rRNAs, and 37 tRNAs. Nineteen genes comprising seven protein-coding genes, four rRNAs, and seven tRNAs are duplicated in the IR regions.
Seven single nucleotide polymorphisms (SNPs) and nine insertion and deletion (INDEL) regions (22 bp in total) were found between A. distichum f. lilacinum and A. distichum ‘Dae Ryun’ (MN116559) using ‘Find Variations/SNPs’ function in Geneious Prime® v2020.2.4 (Biomatters Ltd., Auckland, New Zealand) based on the pair-wise alignment of the two chloroplast genomes. In addition, 6 SNPs and 7 INDEL regions (19 bp) were found against Sang Jae and Ok Hwang 1ho sequences (MN127986 and MK616470) and 99 SNPs and 18 INDEL regions (72 bp) and 6 SNPs and 8 INDEL regions (21 bp) were identified against NC_031445 and MF407183, respectively. These numbers of intraspecific variations except those against NC_031445 are smaller than those identified between Korean isolates of other plant species (Cho et al. Citation2019; Kim et al. Citation2019; Kim et al. Citation2019; Oh et al. Citation2019a, Citation2019b; Park and Kim Citation2019; Park et al. Citation2019a, Citation2019b; Choi et al. Citation2020; Park et al. Citation2021), suggesting that genetic diversity in A. distichum may not be sufficient enough so that morphological features are not tightly linked to those intraspecific variations on chloroplast genomes.
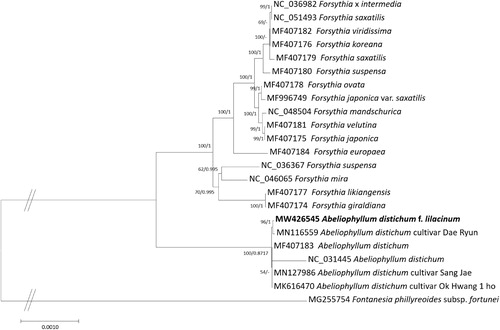
Twenty-three chloroplast genomes representing Abeliophyllum, Forsythia, and Fontanesia genera were used for constructing the bootstrapped Maximum-Likelihood (ML; bootstrap repeat is 1,000) and Bayesian Inference (BI; number of generations is 1,100,000) phylogenic trees using MEGA X (Kumar et al. Citation2018) and MrBayes v3.2.7a (Ronquist et al. Citation2012), respectively, after aligning the whole chloroplast genomes with MAFFT v7.450 (Katoh and Standley Citation2013). ML and BI phylogenetic trees show that A. distichum f. lilacinum is clustered with the Dae Ryun cultivar (MN116559; ) which displays a flower that is bigger than that of the wild type of A. distichum (Park et al. Citation2019). The phylogenetic reconstructions also show that the other A. distichum forma chloroplast genomes are clustered with relatively low supportive values of ML and BI trees (54 and 0.8717, respectively, in ) or are incongruent in both trees (the clade covering NC_031445, MN127986, and MK616470 in ). These analyses indicate that phenotypic variations of A. distichum are not clearly resolved using whole chloroplast genome sequences like the case of Sueda japonica (Kim et al. Citation2020), requiring other genome-wide analysis methods such as Genotyping-By-Sequencing (GBS) which has been applied to various plant species to find relationships between morphological features (Wong et al. Citation2015; Guajardo et al. Citation2020).
Figure 1. Maximum-likelihood (bootstrap repeat is 1,000) and Bayesian Inference (Number of generations is 1,100,000) phylogenetic trees of twenty-three chloroplast genomes of Abeliophyllum, Forsythia, and Fontanesia. The phylogenetic tree was drawn based on maximum-likelihood tree. The numbers above the branches indicate bootstrap support value of maximum likelihood tree and posterior probability of Bayesian Inference phylogenetic tree, respectively.
Disclosure statement
The authors declare that they have no competing interests.
Data availability statement
Chloroplast genome sequence can be accessed via accession number of MW426545 in GenBank of NCBI at https://www.ncbi.nlm.nih.gov. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA689053, SAMN17198782, and SRR13340840, respectively.
Additional information
Funding
References
- Cho M-S, Kim Y, Kim S-C, Park J. 2019. The complete chloroplast genome of Korean Pyrus ussuriensis Maxim. (Rosaceae): providing genetic background of two types of P. ussuriensis. Mitochondrial DNA Part B. 4(2):2424–2425.
- Choi YG, Yun N, Park J, Xi H, Min J, Kim Y, Oh S-H. 2020. The second complete chloroplast genome sequence of the Viburnum erosum (Adoxaceae) showed a low level of intra-species variations. Mitochondrial DNA Part B. 5(1):271–272.
- Guajardo V, Solís S, Almada R, Saski C, Gasic K, Moreno MÁ. 2020. Genome-wide SNP identification in Prunus rootstocks germplasm collections using Genotyping-by-Sequencing: phylogenetic analysis, distribution of SNPs and prediction of their effect on gene function. Sci Rep. 10(1):1–14.
- Jang TW, Choi JS, Park JH. 2020. Protective and inhibitory effects of acteoside from Abeliophyllum distichum Nakai against oxidative DNA damage. Mol Med Rep. 22(3):2076–2084.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kim Y, Heo K-I, Park J. 2019. The second complete chloroplast genome sequence of Pseudostellaria palibiniana (Takeda) Ohwi (Caryophyllaceae): intraspecies variations based on geographical distribution. Mitochondrial DNA Part B. 4(1):1310–1311.
- Kim Y, Park J, Chung Y. 2020. The comparison of the complete chloroplast genome of Suaeda japonica Makino presenting different external morphology (Amaranthaceae). Mitochondrial DNA Part B. 5(2):1616–1618.
- Kim Y, Yi J-S, Min J, Xi H, Kim DY, Son J, Park J, Jeon J-I. 2019. The complete chloroplast genome of Aconitum coreanum (H. Lév.) Rapaics (Ranunculaceae). Mitochondrial DNA Part B. 4(2):3404–3406.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Lee TB. 1976. New forms of Abeliophyllum distichum. Korean J Pl Taxon. 7(1):21–22.
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv preprint arXiv:13033997.
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The sequence alignment/map format and SAMtools. Bioinformatics. 25(16):2078–2079.
- Melchior H. 1964. Syllabus der pflanzenfamilien II. Berlin: Gebrüder Borntraeger.
- Min J, Kim Y, Xi H, Jang T, Kim G, Park J, Park J-H. 2019. The complete chloroplast genome of a new candidate cultivar, Sang Jae, of Abeliophyllum distichum Nakai (Oleaceae): initial step of A. distichum intraspecies variations atlas. Mitochondrial DNA Part B. 4(2):3716–3718.
- Nakai T. 1919. Genus novum Oleacearum in Corea media inventum. Shokubutsugaku Zasshi. 33(392):153–154.
- Oh H, Kang DG, Kwon TO, Jang KK, Chai KY, Yun YG, Chung HT, Lee HS. 2003. Four glycosides from the leaves of Abeliophyllum distichum with inhibitory effects on angiotensin converting enzyme. Phytother Res. 17(7):811–813.
- Oh S-H, Suh HJ, Park J, Kim Y, Kim S. 2019a. The complete chloroplast genome sequence of a morphotype of Goodyera schlechtendaliana (Orchidaceae) with the column appendages. Mitochondrial DNA Part B. 4(1):626–627.
- Oh S-H, Suh HJ, Park J, Kim Y, Kim S. 2019b. The complete chloroplast genome sequence of Goodyera schlechtendaliana in Korea (Orchidaceae). Mitochondrial DNA Part B. 4(2):2692–2693.
- Park J, An J-H, Kim Y, Kim D, Yang B-G, Kim T. 2020. Database of National Species List of Korea: the taxonomical systematics platform for managing scientific names of Korean native species. J Species Res. 9(3):233–246.
- Park J, Kim Y. 2019. The second complete chloroplast genome of Dysphania pumilio (R. Br.) mosyakin & clemants (Amranthaceae): intraspecies variation of invasive weeds. Mitochondrial DNA Part B. 4(1):1428–1429.
- Park J, Kim Y, Xi H, Jang T, Park J-H. 2019. The complete chloroplast genome of Abeliophyllum distichum Nakai (Oleaceae), cultivar Ok Hwang 1ho: insights of cultivar specific variations of A. distichum. Mitochondrial DNA Part B. 4(1):1640–1642.
- Park J, Kim Y, Xi H, Oh YJ, Hahm KM, Ko J. 2019a. The complete chloroplast genome of common camellia tree in Jeju island, Korea, Camellia japonica L. (Theaceae): intraspecies variations on common camellia chloroplast genomes. Mitochondrial DNA Part B. 4(1):1292–1293.
- Park J, Kim Y, Xi H, Oh YJ, Hahm KM, Ko J. 2019b. The complete chloroplast genome of common camellia tree, Camellia japonica L.(Theaceae), adapted to cold environment in Korea. Mitochondrial DNA Part B. 4(1):1038–1040.
- Park J, Min J, Kim Y, Xi H, Kwon W, Jang T, Kim G, Park J-H. 2019. The complete chloroplast genome of a new candidate cultivar, Dae Ryun, of Abeliophyllum distichum Nakai (Oleaceae). Mitochondrial DNA Part B. 4(2):3713–3715.
- Park J, Xi H, Han J, Lee J, Kim Y, Lee J-m, Son J, Ahn J, Jang T, Choi J. 2020. Prediction and identification of biochemical pathway of acteoside from whole genome sequences of Abeliophyllum Distichum Nakai, cultivar Ok Hwang 1ho. J Converg Inf Technol. 10(3):76–91.
- Park, J., Min, J., Kim, Y. and Chung, Y., 2021. The Comparative Analyses of Six Complete Chloroplast Genomes of Morphologically Diverse Chenopodium album L. (Amaranthaceae) Collected in Korea. International Journal of Genomics, 2021.
- Ronquist F, Teslenko M, Van Der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542.
- Wong MM, Gujaria-Verma N, Ramsay L, Yuan HY, Caron C, Diapari M, Vandenberg A, Bett KE. 2015. Classification and characterization of species within the genus Lens using genotyping-by-sequencing (GBS). PLoS One. 10(3):e0122025.
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18(5):821–829.
- Zhao Q-Y, Wang Y, Kong Y-M, Luo D, Li X, Hao P. 2011. Optimizing de novo transcriptome assembly from short-read RNA-Seq data: a comparative study. BMC Bioinf. 12(Suppl 14):S2.
