Abstract
Grewia biloba is a potential medicinal and ornamental resource, and this study is the first to assemble the chloroplast genome of this species using high-throughput sequencing data. The chloroplast genome of G. biloba has a typical composition for higher plant chloroplasts, containing one large single-copy region of 86,978 bp and one small single-copy region of 20,140 bp, which are separated by a pair of inverted-repeat regions of 25,473 bp. The chloroplast genome of this species encodes 130 genes, including 84 protein-coding genes, 38 tRNA genes and 8 rRNA genes. A maximum-likelihood phylogenetic tree was constructed based on sequence information of the chloroplast genomes. The chloroplast genome of G. biloba will provide valuable genetic information for evolutionary research and utilization of this species.
Grewia biloba G. Don is a deciduous shrub or small tree of the genus Grewia in the family Malvaceae. This species is primarily distributed in southern China, with a small number observed in northern China. In some Asian countries and regions, several plants of this genus are used as folk medicinal herbs and are occasionally cultivated in gardens as ornamental plants (Yang and Gao Citation2013). A recent pharmacological study has shown that ethanol extracts of G. biloba root bark and solvent extracts with different polarities had antitumor effects on cervical cancer growth in mice (Liu et al. Citation2008). An n-butanol extract from G. biloba branches showed antibacterial, antimalarial and anti-inflammatory effects (Yang and Gao Citation2013). In addition, an ethyl acetate fraction of G. biloba showed analgesic effects, which might be related to the inhibition of prostaglandin E2 and malondialdehyde, as well as the promotion of nitric oxide release (Yang et al. Citation2014). In this study, we assembled the chloroplast (cp) genome of G. biloba using high-throughput sequencing data and constructed a phylogenetic tree based on cp genome information of related species, which may help establish a foundation for evolutionary research, development and utilization of this species.
Fresh leaf samples of G. biloba were collected on the campus of Henan Agricultural University (34°47′7″N, 113°39′35″E), Zhengzhou city, China. The specimens (MALGR20190815) were preserved in the herbarium of Henan Agricultural University. Total genomic DNA was extracted using a modified cetyltrimethylammonium bromide method (Fang et al. Citation2009). The purified DNA was subsequently fragmented and used to construct short-insert libraries according to the manufacturer’s instructions (Illumina) and then sequenced on an Illumina HiSeq 4000 system (Borgstrom et al. Citation2011). After the raw reads were filtered, the clean reads were assembled into circular genomes using GetOrganelle (Jin et al. Citation2018). Annotations and adjustments of genes were conducted manually using Geneious Prime (Kearse et al. Citation2012). The complete cp genome of G. biloba was submitted to GenBank (accession number: MW699774).
The cp genome of G. biloba is 158,064 bp in length and has a typical structure of a higher plant cp genome; that is, it contains one large single-copy region and one small single-copy region, which are separated by a pair of inverted-repeat regions. The sizes of the above three regions are 86,978, 20,140 and 25,473 bp, respectively. The cp genome of this species encodes 130 genes, including 84 protein-coding genes, 38 tRNA genes and 8 rRNA genes. The GC content of the whole genome is 37.4%, which is similar to that of other species in this family (Alzahrani Citation2021; Wang et al. Citation2021).
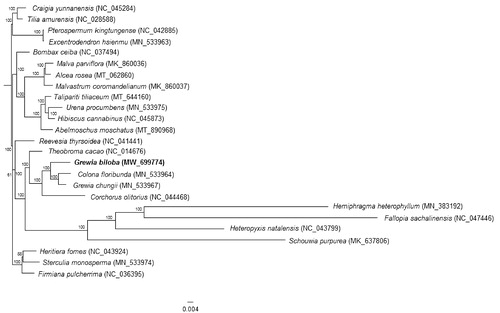
The assembled cp genome of G. biloba was aligned with the cp genomes of 24 other Malvaceae species, which were downloaded from the NCBI database using the MAFFT method in Geneious Prime (Kearse et al. Citation2012). A maximum-likelihood (ML) phylogenetic tree was subsequently constructed using RAxML-HPC2 on XSEDE v8.2.12 on the CIPRES cluster (Miller et al. Citation2010). In the constructed phylogenetic tree, Colona floribunda grouped with another species of the genus Grewia, G. chungii, and formed a sister clade with G. biloba (). The cp genome of G. biloba may provide valuable genetic information for the study of phylogenetic relationships.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in NCBI GenBank (https://www.ncbi.nlm.nih.gov/nuccore/MW699774). The associated BioProject, SRA, and Bio-Sample numbers are PRJNA707319, SRR13883638, and SAMN18200682, respectively.
References
- Alzahrani DA. 2021. Complete chloroplast genome of Abutilon fruticosum: genome structure, comparative and phylogenetic analysis. Plants. 10(2):270.
- Borgstrom E, Lundin S, Lundeberg J. 2011. Large scale library generation for high throughput sequencing. PLoS One. 6:e19119.
- Fang XM, Wang HW, Cheng YQ, Ye YZ, Yang C. 2009. Optimization of total DNA extraction and test of suitable molecular markers in Taihanggia rupestris. Chin Agricul Sci Bull. 25(18):57–60.
- Jin JJ, Yu WB, Yang JB, Song Y, Yi TS, Li DZ. 2018. GetOrganelle: a simple and fast pipeline for de novo assembly of a complete circular chloroplast genome using genome skimming data. BioRxiv. 2018:1–11.
- Kearse M, Moir R, Wilson A, Stones–Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(2):1647–1649.
- Liu JQ, Wu JM, He ZH, Kou XL, Hong Q. 2008. Stuy on the antibacterial activities of extracts from Grewia biloba G. Don in vitro. Lishizhen Med Materia Medica Res. 19(6):1351–1352.
- Miller MA, Pfeiffer W, Schwartz T. 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. Gateway Computing Environments Workshop (GCE). IEEE. p. 1–8.
- Wang JH, Moore MJ, Wang HX, Zhu ZX, Wang HF. 2021. Plastome evolution and phylogenetic relationships among Malvaceae subfamilies. Gene. 765:145103.
- Yang CG, Gao S. 2013. Study on anti-inflammatory effects and mechanisms of N-butanol extract from Grewia biloba in mince. J Liaoning Normal Univ. 36(4):539–542.
- Yang CG, Gao S, Zhao Y, Li ZX. 2014. Study on the analgesic effects and its mechanisms of ethyl acetate fraction of Grewia biloba in mice. J Liaoning Normal Univ. 37(3):402–405.