Abstract
Anneissia intermedia (A.H. Clark, 1916) is a common crinoid found in waters along the coastlines of China, Japan, and Korea. In this study, we determined the complete mitogenome of A. intermedia. The genome was found to be 15,874 bp in length and consists of 13 protein-coding genes, 22 tRNAs, and 2 rRNAs. With the exception of Antedon mediterranea, the gene order and genetic characteristics of the A. intermedia mitogenome are identical with those of the mitogenomes of other crinoids. The complete mitogenome of A. intermedia will contribute to enhance our understanding of the phylogeny of crinoids.
The genus Anneissia consists of nine species distributed over a wide range of the Indo-western Pacific Ocean, including Australia, New Zealand, New Caledonia, China, Japan, and Korea (Clark and Rowe Citation1971; Shin Citation2013; Messing Citation2020). Although the different species of Anneissia are among the most abundant crinoids in the Pacific region, to date, the sequencing of a complete mitogenome has been reported for only a single species, namely, A. pinguis. In this study, we augment this sole representative of the genus Anneissia by determining the complete mitochondrial genome of A. intermedia. Specimens of A. intermedia, which were identified based on morphological observations, were collected by SCUBA diving at a depth of 14 m in waters off the coast of Busan, Korea, on 25 September 2020 (35.037028 N, 129.091806 E). Voucher specimens and mitochondrial DNA (mtDNA) samples have been deposited at the Marine Echinoderm Resources Bank of Korea (http://www.syu.ac.kr; Taekjun Lee, [email protected]) under the voucher number MERBK-C-00307. mtDNA extraction, amplification, and sequencing were performed in accordance with the methods described by Lee and Shin (Citation2018). The complete mitogenome was annotated using Geneious ver. 11.1.5 (Biomatters Ltd, Auckland, New Zealand), and transfer RNAs (tRNAs) were identified and gene locations determined as described by Lee and Shin (Citation2019). Phylogenetic analysis was performed using 24 complete echinoderm mitogenomes [nine crinoids (including A. intermedia), four asteroids, four ophiuroids, four echinoids, and three holothuroids], with those of two hemichordates (Balanoglossus carnosus and B. clavigerus) being used as an outgroup. Mitogenome sequences were aligned using MAFFT (Katoh and Standley Citation2013) and the mitogenome dataset was analyzed based on the maximum likelihood (ML) method using RAxML 8.2 (Stamatakis Citation2014). The best-fit substitution was estimated using jModelTest 2.1.1 (Guindon and Gascuel Citation2003; Darriba et al. Citation2012) for the nucleotide dataset of 13 protein-coding genes (PCGs). For ML analyses, bootstrap resampling was performed using the rapid option with 1000 iterations.
The complete mitogenome of A. intermedia (GenBank accession No. MW376476) is 15,874 bp in length and contains 13 PCGs, 22 tRNA genes, and two rRNA genes. With the exception of that of Antedon mediterranea, the gene order and direction of PCGs and rRNAs were found to be identical to those of the complete mitogenomes of other comatulid species. All PCGs contain an ATG initiation codon (methionine), and the termination codons are generally either TAA or TAG, with that of cytochrome b, which terminates with (GAG)+T (glutamic acid), being the only exception.
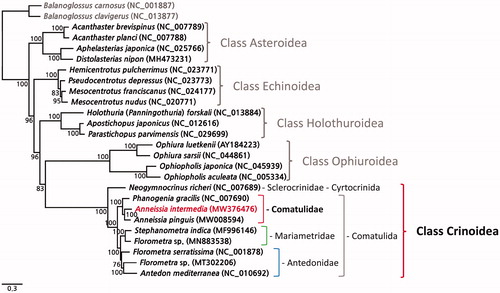
In the constructed phylogenetic tree, the nine complete mitogenomes of crinoids clearly established a monophyletic clade (), within which, A. intermedia clusters with A. pinguis and Phanogenia gracilis, belonging to the same family, Comatulidae (). The sequencing of the complete mitogenome of A. intermedia will provide a basis for further phylogenetic studies on crinoids.
Figure 1. A phylogenetic tree constructed using the maximum likelihood (ML) method based on the nucleotide sequences of 13 protein-coding genes of nine crinoids, including A. intermedia (MW376476), other echinoderms, and two outgroup species (B. carnosus and B. clavigerus). Bootstrap support values were indicated on each node.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are available at https://www.ncbi.nlm.nih.gov/, accession number MW376476. The associated BioProject, Bio-Sample, and SRA numbers are PRJNA715061, SAMN18325186, and SRR13996249, respectively.
Additional information
Funding
References
- Clark AM, Rowe FWE. 1971. Monograph of shallow-water Indo-West Pacific echinoderms. London: Trustees of the British Museum of Natural History.
- Darriba D, Taboada GL, Doallo R, Posada D. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 9(8):772.
- Guindon S, Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 52(5):696–704.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Lee T, Shin S. 2018. Complete mitochondrial genome analysis of Distolasterias nipon (Echinodermata, Asteroidea, Forcipulatida). Mitochondrial DNA Part B. 3(2):1290–1291.
- Lee T, Shin S. 2019. Complete mitochondrial genome of sea cucumber, Holothuria (Stauropora) pervicax (Holothuroidea, Holothuriida, Holothuriidae), from Jeju Island, Korea. Mitochondrial DNA Part B. 4(1):1047–1048.
- Messing C. 2020. World list of Crinoidea. Anneissia Summers, Messing & Rouse, 2014. World Register of Marine Species; [accessed 2020 Dec 24]. http://www.marinespecies.org/aphia.php?p=taxdetails&id=828991on
- Shin S. 2013. Echinodermata: Crinozoa: Crinoidea: Comatulida, Asterozoa, Ophiuroidea: Euryalida feather stars, Basket Stars. Invertebrate fauna of Korea. 5th ed. Vol. 32. Incheon: National Institute of Biological Resources.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
