Abstract
Sinocyclocheilus longibarbarus and Sinocyclocheilus punctatus were collected from a karst cave Libo County, southwest of China. The two Sinocyclocheilus species can be distinguished obviously by external morphological characteristics. In this study, the complete mitochondrial genome sequences of two species were assembled, and both sequences reflected gene organization typical for mitochondrial DNA of the genus Sinocyclocheilus, comprising of 13 protein-coding genes (PCGs), 22 transfer RNA genes (tRNAs), 2 ribosomal RNA genes (rRNAs), and a large non-coding control region. Phylogenetic analysis showed that S. punctatus was first clustered together with S. mutipunctatus, and S. longibarbarus was closely related to S. yishanensis. The complete mitogenome of two species may provide useful information for the further taxonomic and phylogenetic studies.
Sinocyclocheilus genus is an endemic group in China; due to the isolation of cave environment, the species within the genus are strongly differentiated. To date, 75 species of effective species have been recorded, and most of the species live in caves and underground rivers. It is widely distributed in karst areas such as Yunnan, Guizhou and Guangxi. Sinocyclocheilus longibarbarus is only distributed in Guizhou (Wang and Chen 1989), while Sinocyclocheilus punctatus is distributed in Nandan, Huanjiang and Libo areas (Lan et al. Citation2017). Both of them belong to semi cave species with normal eyes and covered with scales, meanwhile, the two species can be distinguished by external morphological characteristics such as angular whisker, body shape, scale and body spot. Interestingly, the two species were collected from a karst cave in Libo County (23°14′15″N, 108°02′18″E), Guizhou Province, belonging to the Liujiang River System in the Pearl River Basin, and the specimen were fixed in 95% ethanol and preserved in Liuzhou Aquaculture Technology Extending Station, Liuzhou, China. Total genomic DNA were extracted from the fins through using traditional phenol-chloroform extraction method (Taggart et al. Citation1992). Then, we sequenced the complete mitochondrial genomes of the two Sinocyclocheilus species using the Illumina Hiseq4000 platform with de novo strategy (Tang et al. Citation2015) and submitted the genomes to GenBank with accession numbers MT361975 (S. longibarbarus) and MT361976 (S. punctatus), respectively.
The length of two complete circular mitogenomes were 16,570 bp with 44.08% GC content (S. longibarbarus) and 16,582 bp with 43.60% GC content (S. punctatus) respectively. Both genomes consisted of 13 protein-coding genes, 22 transfer RNAs, and 2 ribosomal RNAs, and the arrangement of all genes were identical to other Sinocyclocheilus species known to us (Wu et al. Citation2010; He et al. Citation2016; Luo et al. Citation2017; Peng et al. Citation2017). All genes of two Sinocyclocheilus species were located on the heavy strand (H-strand) except for ND6 gene and eight tRNA genes, and the protein-coding genes started with a traditional ATG except for COX1, which started with one infrequent GTG instead, and terminated with stop codons TAA, TAG, or a single T-base. The control region (D-loop) of two Sinocyclocheilus species were located between tRNA-Phe and tRNA-Pro, with 920 bp (S. longibarbarus) and 928 bp in length (S. punctatus), respectively.
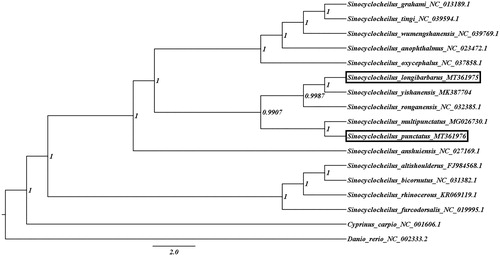
To determine the phylogenetic relationship of both species in the Sinocyclocheilus family, a Bayesian Inference (BI) tree was constructed on MrBayes v3.2.7 using the 13 PCGs in mitogenomes of 17 species. Danio rerio and Cyprinus carpio was set as outgroups. We obtained the best-fit substiution model (GTR + I + G) by MrModelTest 2.3 (Nylander Citation2004) in PAUP 4.0, and Four parallel runs of Markov Chain Monte Carlo (MCMC) were run for 1,000,000 generations, sampling every 1000 generations and discarded 100 trees as burn-in. Phylogenetic analysis showed that S. punctatus was not first clustered together with S. longibarbarus which lived in the same cave but S. mutipunctatus, and S. longibarbarus was closely related to S. yishanensis (), it suggested that long-term reproductive isolation might lead to species differentiation, while convergent evolution had little effect on species differentiation in similar environments.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov, reference number MT361975 (Sinocyclocheilus longibarbarus) and MT361976 (Sinocyclocheilus longibarbarus).
Additional information
Funding
References
- He SY, Lu J, Jiang WS, Yang S, Yang JX, Shi Q. 2016. The complete mitochondrial genome sequence of a cavefish Sinocyclocheilus anshuiensis (Cypriniformes: Cyprinidae). Mitochondrial DNA. 27:4256–4258.
- Lan YB, Qin XC, Lan JH, Xiu LH, Yang J. 2017. A new species of the genus (Cypriniformes,Cyprinidae) from Guangxi, China. J Xinyang Normal Univ (Nat Sci Ed). 30(1):97–101. (in Chinese with English abstracts).
- Luo FG, Huang J, He AY, Luo T, Zhou HJ, Wen YH. 2017. Complete mitochondrial genome of a cavefish Sinocyclocheilus ronganensis (Cypriniformes: Cyprinidae). Mitochondrial DNA Part B. 2:117–118.
- Nylander JAA. 2004. MrModeltest v2. Program distributed by the author. Uppsala: Evolutionary Biology Centre, Uppsala University.
- Peng S, Lu J, Yang JX, Chen XL, Shi Q. 2017. The complete mitochondrial genome of horned Golden-line barbell, Sinocyclocheilus rhinocerous (Cypriniformes, Cyprinidae). Mitochondrial DNA. 28:269–270.
- Taggart J, Hynes R, Prod Uhl P, Ferguson A. 1992. A simplified protocol for routine total DNA isolation from salmonid fishes. J Fish Biol. 40(6):963–965.
- Tang M, Hardman CJ, Ji Y, Meng G, Liu S, Tan M, Yang S, Moss ED, Wang J, Yang C, et al. 2015. High-throughput monitoring of wild bee diversity and abundance via mitogenomics. Methods Ecol Evol. 6:1034–1043.
- Wang DZ, Chen YY. 1989. Ddscriptions of three new species of Cyprinidae from Guizhou Province, China (Cypriniformcs: Cyprinidae). Acta Acad Med Zunyi. 12(4):29–34. (in Chinese with English abstracts).
- Wu XJ, Wang L, Chen SY, Zan RG, Xiao H, Zhang YP. 2010. The complete mitochondrial genomes of two species from Sinocyclocheilus (Cypriniformes: Cyprinidae) and a phylogenetic analysis within Cyprininae. Mol Biol Rep. 37:2163–2171.