Abstract
Thyrsostachys is oligotypic genus of Bambusinae, while its phylogenetic position had been unclear. Here, the complete plastid genome of the type species, T. siamensis, was sequenced and analyzed in this work. The complete genome is a typical quadripartite structure with 139,522 bp in length, comprising of a large single-copy region (LSC, 83,032 bp), a small single-copy region (SSC, 12,892 bp), and a pair of invert repeats regions (IR, 21,799 bp). The genome contains 138 genes, 89 protein-coding genes, 41 tRNA genes, and 8 rRNA genes. The GC content of genome was 38.9%. Phylogenetic analysis indicated T. siamensis was sister to Dendrocalamus birmanicus within Bambusinae.
Keywords:
The Bambusoideae is one of the largest subfamilies of the grass family (Poaceae) with approximately 1680 species (Liu et al. Citation2020). The phylogenetic classification of this subfamily recognizes three tribes, Arundinarieae, Olyreae and Bambuseae (Soreng et al. Citation2017). Among them, the Bambuseae is the largest tribe with approximately 970 species and divided into two major lineages: the neotropical woody bamboos (including Arthrostylidiinae, Chusqueinae and Guaduinae) and the paleotropical woody bamboos (including Bambusinae, Dinochloinae, Greslaniinae, Hickeliinae, Holttumochloinae, Melocanninae, Temburongiinae and Racemobambosinae) (Soreng et al. Citation2017; Zhou et al. Citation2017).
Thyrsostachys Gamble, belonging to Bambusinae, includes only two species, T. siamensis and T. oliveri, which are native to Yunnan, Myanmar, Thailand, Vietnam, and distributed from tropics to temperate regions of Asia (Li et al. Citation2006). Previous studies suggested that the position of Thyrsostachys was not clear. Based on the ITS, GBSSI and trnL-F sequences, the phylogenetic analysis of paleotropical woody bamboos showed that Thyrsostachys was monophyletic and sister to Dendrocalamus, Gigantochloa, Oxytenanthera, and Neosinocalamus (Yang et al. Citation2008). Based on the GBSSI, psbA-trnH, rpl32-trnL and rps16 intron sequences, the phylogenetic analysis of Bambusa and its allies showed that Thyrsostachys was sister to Melocalamus, Dendrocalamus, Gigantochloa, and Oxytenanthera (Yang et al. Citation2010). Present study sequenced and assembled a complete plastid genome of the type species T. siamensis to investigate the genome feature and phylogenetic position.
The sample was acquired from Cangshan District, Fujian Province of China (26°38′N, 119°27′E), and the voucher specimen deposited in the Herbarium of the Forestry College of Fujian Agriculture and Forestry University (FJFC, specimen code Bamboo0910). Complete plastid genome of Dendrocalamus latiflorus (FJ970916) as reference, the paired-end reads were filtered by GetOrganelle pipe-line (https://github.com/Kinggerm/GetOrganelle) to get plastid-like reads (Jin et al. Citation2020), then the filtered reads were assembled by SPAdes version 3.10 (Bankevich et al. Citation2012). Then the final ‘fastg’ were filtered by the script of GetOrganelle to get pure plastid contigs, and the filtered De Brujin graphs were viewed and edited by Bandage (Wick et al. Citation2015). Assembled plastid genome annotation based on comparison with D. latiflorus by GENEIOUS v11.1.5 (Biomatters Ltd., Auckland, New Zealand) (Kearse et al. Citation2012). The matrix of 60 representative species of paleotropical woody bamboos were aligned using MAFFT v7.307 (Katoh and Standley Citation2013). Five genera of neotropical woody bamboos (Chusquea circinata of Chusqueinae; Guadua angustifolia, Otatea glauca and O. reflexa of Guaduinae; and Merostachys sp. and Rhipidocladum pittieri of Arthrostylidiinae) were selected as the outgroup according to Zhou et al. (Citation2017) and Liu et al. (Citation2020). The phylogenetic tree was constructed based on the complete plastid genomes by the maximum likelihood software IQ-TREE (Nguyen et al. Citation2015) and branch supports with the ultrafast bootstrap (Hoang et al. Citation2018). The analyses were performed using Phylosuite platform (Zhang et al. Citation2020).
The complete plastid genome of T. siamensis was a quadripartite structure with the length of 139,522 bp, contains a large single copy region (LSC) of 83,032 bp, a small single copy (SSC) region of 12,892 bp, and two inverted repeat (IR) regions of 21,799 bp. The genome possessed total 139 genes, including 89 protein-coding genes, 8 rRNA genes, and 41 tRNA genes. The overall GC content of the plastid genome was 38.9%, while the corresponding values of the LSC, SSC, and IR regions are 37.0%, 33.1%, and 44.2%, respectively.
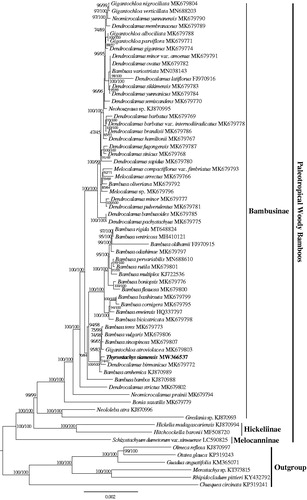
The phylogenetic analysis of 65 Bambuseae plastomes showed that the paleotropical woody bamboos were grouped into three major clades, Melocanninae, Hickeliinae, and Bambusinae, with high support (). The T. siamensis was sister to Dendrocalamus birmanicus within the Bambusinae clade. This newly reported plastid genome provides a good foundation for better understanding the generic relationships of subtribe Bambusinae.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The complete plastid genome of Thyrsostachys siamensis of this study is available in NCBI GenBank database (https://www.ncbi.nlm.nih.gov) with the accession code MW366537.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS. 2018. UFBoot2: improving the ultrafast bootstrap approximation. Mol Biol Evol. 35:518–522.
- Jin JJ, Yu WB, Yang JB, Song Y, de Pamphilis CW, Yi TS, Li DZ. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genom Biol. 21:241.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Li DZ, Wang ZP, Zhu ZD, Xia NH, Jia LZ, Guo ZH, Yang GY, Stapleton CMA. 2006. Bambuseae (Poaceae). In: Wu ZY, Raven PH, Hong DY, editors. Flora of China, vol. 22. Beijing and St. Louis: Science Press and Missouri Botanical Garden Press.
- Liu JX, Zhou MY, Yang GQ, Zhang YX, Ma PF, Guo C, Vorontsovae MS, Li DZ. 2020. ddRAD analyses reveal a credible phylogenetic relationship of the four main genera of Bambusa-Dendrocalamus-Gigantochloa complex (Poaceae: Bambusoideae). Mol Phylogenet Evol. 146:106758.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-Likelihood phylogenies. Mol Biol Evol. 32:268–274.
- Soreng RJ, Peterson PM, Romaschenko K, Davidse G, Teisher KJ, Clark LJ, Barber P, Gillespie LJ, Zuloaga FO. 2017. A worldwide phylogenetic classification of the Poaceae (Gramineae) II: an update and a comparison of two 2015 classifications. J Syst Evol. 55(4):259–290.
- Wick RR, Schultz MB, Zobel J, Holt KE. 2015. Bandage: interactive visualization of de novo genome assemblies. Bioinformatics. 31(20):3350–3352.
- Yang HQ, Yang JB, Peng ZH, Gao J, Yang YM, Peng S, Li DZ. 2008. A molecular phylogenetic and fruit evolutionary analysis of the major groups of the paleotropical woody bamboos (Gramineae: Bambusoideae) based on nuclear ITS, GBSSI gene and plastid trnL-F DNA sequences. Mol Phylogenet Evol. 48:809–824.
- Yang JB, Yang HQ, Li DZ, Wong KM, Yang YM. 2010. Phylogeny of Bambusa and its allies (Poaceae: Bambusoideae) inferred from nuclear GBSSI gene and plastid psbA-trnH, rpl32-trnL and rps16 intron DNA sequences. Taxon. 59(4):1102–1110.
- Zhang D, Gao FL, Jakovlić I, Li WX, Zhou H, Zhang J, Wang GT. 2020. PhyloSuite: an integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol Ecol Resour. 20(1):348–355.
- Zhou MY, Zhang YX, Haevermans T, Li DZ. 2017. Towards a complete generic-level plastid phylogeny of the paleotropical woody bamboos (Poaceae: Bambusoideae). Taxon. 66(3):539–553.