Abstract
Centranthera grandiflora is an important medicinal herb within Orobanchaceae. To date, however, genetic studies on this species remain poor. Here, we assembled the complete chloroplast genome of C. grandiflora. Results showed that the genome was 147 655 bp in length, consisting of large and small single copy regions of length 83 550 and 14 891 bp, respectively, separated by two inverted repeat regions of 24 607 bp. Furthermore, the genome contained 132 genes, including 84 protein-coding genes, 39 tRNA genes, and eight rRNA genes. Phylogenetic analysis showed that C. grandiflora is closely related to the species of Orobanchaceae. The complete chloroplast genome of C. grandiflora should help in the conservation of genetic resources and appropriate utilization of this medicinal herb in the future.
Centranthera grandiflora Benth is a medicinal herb and is administered to promote blood circulation and reduce blood stasis (Zhu Citation2012). However, only few studies have isolated and identified chemical compounds from this species (Liao et al. Citation2012; Hu Citation2016) and limited molecular studies have only been carried out recently (Zhang et al. Citation2019). Therefore, to facilitate genetic studies in the future, we assembled the complete chloroplast genome of C. grandiflora in this study.
Fresh leaves were collected from Pingbian, Yunnan, China (103°39′17″E, 23°03′16″N). The specimen was stored at Yunnan University of Chinese Medicine (specimen code YUCM2019044). Total genomic DNA was extracted using a Tiangen Plant Kit (Beijing, China). A purified genomic DNA library was constructed and sequenced using the Illumina NovaSeq 6000 Platform (Benagen Tech Solution Co., Ltd, Wuhan, China). Genome was assembled using SPAdes v3.6.1 (Bankevich et al. Citation2012). The annotation was performed with Plastid Genome Annotator (PGA) (Qu et al. Citation2019).
The complete genome of C. grandiflora was 147 655 bp in length, consisting of large and small single copy regions of length 83 550 and 14 891 bp, respectively, separated by two inverted repeat regions of 24 607 bp. The complete chloroplast genome of C. grandiflora displayed the typical quadripartite structure of most angiosperm chloroplast genomes. The overall GC content was 38.24%. Furthermore, the genome contained 132 genes, including 84 protein-coding genes, 39 tRNA genes, and eight rRNA genes.
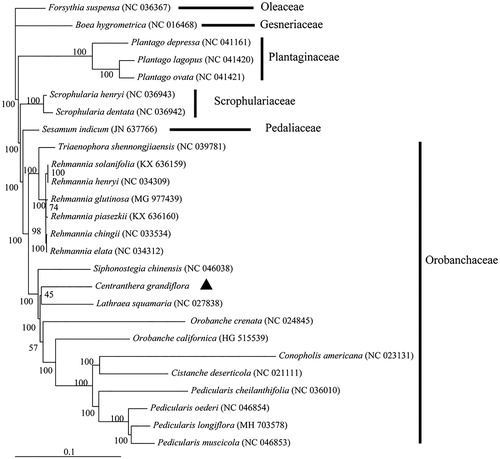
Phylogenetic analysis was performed based on the complete chloroplast genome of C. grandiflora and 25 related species (). MAFFT v7 was used to align the sequences (Katoh and Standley Citation2013), and RAxML v8.2.10 was used to construct a maximum-likelihood (ML) tree (Stamatakis Citation2014). Results showed that C. grandiflora formed a clade with species within Orobanchaceae with 100% support (). Centranthera grandiflora was originally classified in Scrophulariaceae in the Flora of China (Editorial Committee of Flora of Chinese Academy of Sciences Citation1990). With the split of the Scrophulariaceae following the molecular analyses, Centranthera was classified into the Orobanchaceae (Chen et al. Citation2016). Our phylogenetic analysis confirms this placement. The complete chloroplast genome of C. grandiflora should help in the conservation of genetic resources and appropriate utilization of this medicinal herb in the future.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. MW262988. The associated BioProject number is PRJNA706067.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich A, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Chen Z-D, Yang T, Lin L, Lu L-M, Li H-L, Sun M, Liu B, Chen M, Niu Y-T, Ye J-F, et al. 2016. Tree of life for the genera of Chinese vascular plants. J Syst Evol. 54(4):277–306.
- Editorial Committee of Flora of Chinese Academy of Sciences. 1990. Flora of China, Scrophulariaceae. Vol 67. Beijing: Science Press.
- Hu Q. 2016. A study on chemical composition of Centranthera grandiflora. Clin J Chin Med. 8(22):49–53.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Liao L, Zhang Z, Hu Z, Chou G, Wang Z. 2012. Iridoid glycosides from Centranthera grandiflora. Chin Tradit Herbal Drugs. 43(12):2369–2371.
- Qu X-J, Moore MJ, Li D-Z, Yi T-S. 2019. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods. 15:50.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Zhang X, Li C, Wang L, Fei Y, Qin W. 2019. Analysis of Centranthera grandiflora benth transcriptome explores genes of catalpol, acteoside and azafrin biosynthesis. Int J Mol Sci. 20(23):6034.
- Zhu Z. 2012. Illustrated handbook for medicinal materials from nature in Yunnan. Vol 5. Kunming: Yunnan Science and Technology Press.