Abstract
Lonicera tatarica L. is an excellent landscaping shrub with high ornamental value. Here, we report the complete chloroplast genome sequence of L. tatarica. The size of the chloroplast genome is 154,675 bp in length, including a large single copy region (LSC) of 88,361 bp, a small single copy region (SSC) of 18,750 bp, and a pair of inverted repeated regions of 23,782 bp. The L. tatarica chloroplast genome encodes 131 genes, including 85 protein-coding, 38 tRNA, and 8 rRNA genes. Phylogenetic analysis fully resolved L. tatarica in a clade with L. japonica, L. confusa, and L. maximowiczii. These data provide a useful resource when studying the genetic diversity of L. tatarica.
The genus Lonicera is classified in Caprifoliaceae and comprises more than 200 species (Naugžemys et al. Citation2007). Some of the taxa assigned to in this genus are used to facilitate erosion control as well as for ornamental purposes. One of them, Lonicera tatarica L., is an excellent landscaping shrub with high ornamental value due to its strong resistance, its ability to easily propagate, and beautiful flowers and leaves (Palacios et al. Citation2002). The emergence of high-throughput sequencing technologies makes it possible to quickly obtain a large amount of genomic data (He et al. Citation2017). Using this technology, this study reports the complete chloroplast (cp) genome of L. tatarica. The raw sequence data have been deposited in NCBI SRA with a project accession of PRJNA687839, and the cp genome sequence has been registered in GenBank with the Accession no. MW340876.
Fresh leaf samples of L. tatarica were collected from the campus of Henan University, China (34°49'19.76"N, 114°18'51.25"E), and a voucher specimen was deposited at the Herbarium of Henan University (Voucher no. HENU20200529, Kaifeng, Henan Province, China, Wangjun Yuan and [email protected]). The total genomic DNA was extracted using a modified SDS method (Lim et al. Citation2016). The high-quality DNA was fragmented by sonication to a size of 350 bp. The paired-end libraries were sequenced using Illumina NovaSeq PE150 at Beijing Novogene Bioinformatics Technology Co., Ltd (Beijing, China). This yielded about 5G of raw data. Low-quality bases (mass value ≤20) over a certain percentage (the default was 40%) were removed, and some bases the overlap between them and the Adapter exceeding a certain threshold (the default was 15 bp) and less than 3 mismatches between them were thrown out. Approximately 2.5 Gb clean data were obtained, which were trimmed and assembled into the complete cp genome of L. tatarica using a CLC Genomics Workbench9.5.2 (CLC Inc., Rarhus, Denmark) with the sequence of L. maackii (GenBank Accession no. MN256451) as a reference. Then the cp genome was annotated using Geneious R11 (Biomatters, Auckland, New Zealand) while following the description provided by Liu et al. (Citation2017). The phylogenetic position of L. tatarica was inferred using the whole cp genome sequences, and the maximum-likelihood (ML) method with nucleotide substitution model GTR + G and was implemented in RAxML-HPC v8.1.11 on the CIPRES cluster (Stamatakis Citation2014).
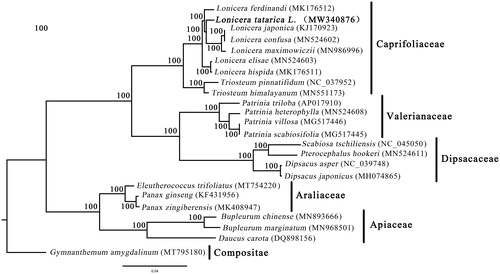
The complete cp genome of L. tatarica possesses a typical quadripartite structure with a length of 154,675 bp, which includes two inverted repeat (IR) regions of 23,782 bp, a large single-copy (LSC) region of 88,361 bp, and a small single-copy (SSC) region of 18,750 bp. The genome contains a total of 131 genes (85 protein-coding genes, 8 ribosomal RNA genes, and 38 tRNA genes). Among these genes, nine protein-coding genes (atpF, ndhA, rpoc1, ndhB, petD, rpl16, rpl2, rps12, and rps16) contain one intron, whereas two genes (clpP and ycf3) contain two introns. Notably, the rps12 gene was trans-spliced with the 5′ end located in the LSC region and the 3′ end duplicated in the IR region. Additionally, the overall GC content was 38.4%, and the corresponding values of the LSC, SSC, and IR regions were 34.2%, 30.0%, and 42.5%, respectively. Furthermore, the phylogenic assessment revealed that L. tatarica was closely clustered with L. japonica, L. confusa, and L. maximowiczii with high bootstrap support values (), and the topology of Caprifoliaceae, Valerianaceae and Dipsacaceae is identical to that of Xiang et al. (Citation2020). These data provide a useful resource when studying the genetic diversity of L. tatarica.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the Accession no. MW340876. The associated ﹡BioProject﹡, ﹡SRA﹡, and﹡Bio-Sample﹡ numbers of the raw sequence data and the genome are PRJNA687839, SRP300309, and SAMN17156276, respectively.
Additional information
Funding
References
- He YX, Dong MF, Yuan WJ, Shang FD. 2017. The first genetic map in sweet osmanthus (Osmanthus fragrans lour.) using specific locus amplified fragment sequencing. Front Plant Sci. 8:1621.
- Lim HJ, Lee EH, Yoon Y, Chua B, Son A. 2016. Portable lysis apparatus for rapid single-step DNA extraction of Bacillus subtilis. J Appl Microbiol. 120(2):379–387.
- Liu LX, Li R, Worth JRP, Li X, Li P, Cameron KM, Fu CX. 2017. The complete chloroplast genome of Chinese bayberry (Morella rubra, Myricaceae): implications for understanding the evolution of Fagales. Front Plant Sci. 8:968.
- Naugžemys D, Žilinskaitė S, Denkovskij J, Patamsytė J, Literskis J, Žvingila D. 2007. RAPD based study of genetic variation and relationships among Lonicera germplasm accessions. Biologija. 53:34–39.
- Palacios N, Christou P, Leech M. 2002. Regeneration of Lonicera tatarica plants via adventitious organogenesis from cultured stem explants. Plant Cell Rep. 20(9):808–813.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Xiang C‐L, Dong H‐J, Landrein S, Zhao F, Yu W‐B, Soltis DE, Soltis PS, Backlund A, Wang H‐F, Li D‐Z, et al. 2020. Revisiting the phylogeny of Dipsacales: new insights from phylogenomic analyses of complete plastomic sequences. J Syst Evol. 58(2):103–117.