Abstract
Strobilanthes tonkinensis Lindau is a member of the family Acanthaceae, which was originated from Yunnan province of China and is used as tea and health promotion. Here, we reported the complete chloroplast genome sequence of S. tonkinensis using Illumina high-throughput sequencing approach. The size of the chloroplast genome is 144,765 bp in length, containing a pair of inverted repeats (IRs, 17,362 bp) that are separated by the large single-copy (LSC, 92,248 bp), and small single-copy (SSC, 17,793 bp) regions. A total of 129 genes were identified, including 37 tRNA genes, 8 rRNA genes, and 84 protein-coding genes. The overall GC content is 38.21%. Phylogenetic analysis indicated that S. tonkinensis is closely related to Strobilanthes cusia and Strobilanthes bantonensis.
Strobilanthes tonkinensis Lindau, 1897, also called Semnostachya menglaensis (Wu et al. Citation2011), is a rare plant of the genus Strobilanthes and a perennial herb of the family Acanthaceae, mainly distributed in Yunnan, China (Srikun Citation2017). S. tonkinensis has been widely used as natural herbal tea and spices in the Southern part of China, which has a distinctive fragrance similar to glutinous rice (Wu et al. Citation2011; Zhang et al. Citation2015). In addition, S. tonkinensis contains a large amount of squalene, which may be responsible for its biological function, such as antioxidative activity (Yang et al. Citation2014; Srikun Citation2017). The complete chloroplast (cp) genome has been widely used as a valuable source of data for understanding evolutionary biology due to its conservation in gene content (Sun et al. Citation2020). In this study, we sequenced and assembled the cp genome of S. tonkinensis for the first time to provide necessary genomic resources for further study and provide important information for the phylogeny of Acanthaceae.
Fresh leaves of S. tonkinensis were collected from Yunnan, China (100°43′51.3804″E, 21°40′55.632 N). The voucher specimen and DNA sample were deposited at the Herbarium of College of pharmaceutical sciences, Zhejiang Chinese Medical University (X. Wang, [email protected]) under the voucher number ZCMU4C505. The total genomic DNA was extracted from S. tonkinensis leaves by a modified CTAB method (Doyle and Doyle Citation1987). After DNA extraction, the genomic DNA was segmented by ultrasound, and a sequencing library with an insert size of 320 bp fragments was constructed by PCR amplification. Then the genomic library was sequenced using the Illumina Novaseq 6000 platform (Illumina, San Diego, CA) in Genepioneer Biotechnologies (Nanjing, China) and approximately 9.11 GB of clean data were obtained. The filtered data were then assembled using NOVOPlasty software (Dierckxsens et al. Citation2016), with Strobilanthes cusia as the reference (GenBank accession number: MG874806.1). Then the assembled sequences of S. tonkinensis were annotated using CpGAVAS (Liu et al. Citation2012) and checked with DOGMA and BLAST (Wyman et al. Citation2004).
The complete chloroplast genome of S. tonkinensis (GenBank accession number: MW525447) is 144,765 bp in length with a typical quadripartite structure containing a pair of inverted repeats (IRs, 17,362 bp), a large single-copy region (LSC, 92,248 bp), and a small single-copy region (SSC, 17,793 bp). The total GC content of the whole genome, IRs, LSC, and SSC were 38.21%, 36.53%, 32.49%, and 45.61%, respectively. A total of 129 genes were successfully annotated, including 8 rRNA genes, 37 tRNA genes, and 84 protein-coding genes. Of which, 16 genes contain one intron, 2 genes contain two introns, and 8 genes were duplicated in the IRs region.
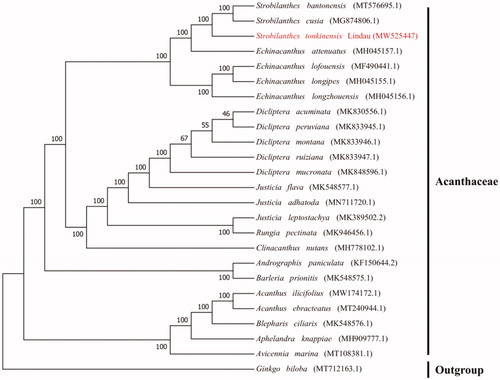
In order to explore the phylogenetic position and evolutionary relationship of S. tonkinensis, a phylogenetic analysis was conducted between S. tonkinensis and other 23 complete chloroplast genomes of the Acanthaceae downloaded from the NCBI Nucleotide Resource database, Ginkgo biloba was used as the outgroup. MAFFT version 7 software (Katoh et al. Citation2005) was used to align the above sequences. A maximum likelihood (ML) tree was built using RA × ML (Alexandros Citation2014) with 1000 bootstrap and with GTRGAMMA as the best nucleotide substitution model. The results indicated that S. tonkinensis belongs to the family Acanthaceae and is closely related to Strobilanthes cusia and Strobilanthes bantonensis ().
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov, under the accession number MW525447. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA727488, SRR14429881, and SAMN19022189, respectively.
Additional information
Funding
Notes on contributors
Liu Fang
Liu Fang is pursuing her master’s degree majoring in Pharmacology under the supervision of Prof. Xingya Wang at the College of Pharmaceutical Sciences, Zhejiang Chinese Medical University of China. Her current research interests are focused on examining the chemical structure and biological functions of Traditional Chinese herbal medicine. E-mail: [email protected].
Ying Wang
Ying Wang is pursuing her Ph.D.’s degree majoring in traditional Chinese medicine under the supervision of Prof. Xingya Wang at the College of Pharmaceutical Sciences, Zhejiang Chinese Medical University of China. Her current research interests are focused on resource development and pharmacological research of natural compounds and Chinese herbal medicine. E-mail: [email protected].
Cui-Ling Guo
Cui-Ling Guo is pursuing her master’s degree in Pharmacology under the supervision of Prof. Xingya Wang at the College of Pharmaceutical Sciences, Zhejiang Chinese Medical University of China. Her current research interests are focused on extracting bioactive compounds from Chinese herbal medicine from different locations and quantitate their amounts. E-mail: [email protected].
Xing-Ya Wang
Xing-Ya Wang, Ph.D., received her master’s and Ph.D.’s diploma from the Ohio State University (from 2001–2007). She then worked at NIHES as a Post-doc fellow from 2007 to 2014. In 2014, she joined Zhejiang Chinese Medical University as a ‘Qianjiang’ distinguished professor in China. Dr. Wang’s work has been focused on examining obesity and its related diseases, including cancer, type 2 diabetes, atherosclerosis, and aging. In addition, Dr. Wang has been interested in studying the resources of Chinese herbal medicines, and their bioactive components isolation and biological function determination. E-mail: [email protected].
References
- Alexandros S. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Dierckxsens N, Mardulyn P, Smits G. 2016. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Katoh K, Kuma K, Toh H, Miyata T. 2005. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 33(2):511–518.
- Liu C, Shi L, Zhu Y, Chen H, Zhang J, Lin X, Guan X. 2012. CpGAVAS, an integrated web server for the annotation, visualization, analysis, and GenBank submission of completely sequenced chloroplast genome sequences. BMC Genom. 13(1):715.
- Srikun N. 2017. In vitro propagation of the aromatic herb Strobilanthes tonkinensis Lindau. Agric Nat Resour. 51(1):15–19.
- Sun J, Wang Y, Liu Y, Xu C, Yuan Q, Guo L, Huang L. 2020. Evolutionary and phylogenetic aspects of the chloroplast genome of Chaenomeles species. Sci Rep. 10(1):1–10.
- Wu G, Ma S, Zhang C, Zhuang F, Zhang J. 2011. Analysis to Genetic in Strobilanthes tonkinensis by RAPD. Chin J Tropical Crops. 32(7):1320–1324.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20(17):3252–3255.
- Yang T, Wu Q, Li S, Lv Z, Hu B, Xie B, Sun Z. 2014. Liposoluble compounds with antioxidant activity from Strobilanthes tonkinensis. Chem Nat Compd. 49(6):1166–1167.
- Zhang Y, Xu F, Tan L, Zhang C, Gu F. 2015. GC-MS analysis of volatiles in Strobilanthes tonkinensis leaf extracted by headspace solid-phase microextraction. Chin J Tropical Crops. 36(3):603–610.