Abstract
The complete mitochondrial genome of the Cerceris quinquefasciata (Rossi, 1792) (Hymenoptera: Crabronidae) was obtained via next-generation sequencing. This mitochondrial genome is 16,188 bp in length with 37 classical eukaryotic mitochondrial genes and two A + T-rich region. All the 13 PCGs begin with typical ATN codons. Among them, eleven PCG genes terminate with TAA, two with T–. All of the 22 tRNA genes, ranging from 58 to 72 bp with typical cloverleaf structure except for trnS1, whose dihydrouridine (DHU) arm forms a simple loop. Phylogenetic analysis highly supported Crabronidae is the sister group of anthophila bees.
Cerceris quinquefasciata (Rossi, 1792) is one of the digger wasp which belongs to genus Cerceris within the family Crabronidae. Five-banded tailed digger wasp is their common name and it is a tiny solitary wasp. They mainly catch small curculionid beetles and paralyze them with their sting (Edwards, Citation1996). As an important solitary wasp, little genetic information is availability. Therefore we sequenced the complete mitochondrial genome of C. quinquefasciata, which was collected by Malaise trap in Weifang University of Science and Technology (36.8915°N, 118.7656°E), Shouguang City in August 2018. The sample used in the experiment are now stored in the Institute of Zoology, Chinese Academy of Sciences (Voucher number: NF20002, E-mail: [email protected]) and the GenBank accession number is MW402864.
The total mitochondrial genome of C. quinquefasciata was obtained through next-generation sequencing. The extracted DNA mixture were applied for library constructing by the usage of Illumina TruSeq@ DNA PCR-Free HT Kit (Illumina, San Diego, CA), and sequenced by the platform of llumina HiSeq sequencer (150 bp pared-end). The mitochondrial genome of C. quinquefasciata was assembled based on Illumina short reads with MitoZ v2.3 (Meng et al. Citation2019). The whole mitochondrial genome annotation was annotated by Mitos WebServer (http://mitos2.bioinf.uni-leipzig.de/index.py) under the invertebrate mitochondrial code (Bernt et al. Citation2013). Transfer RNA (tRNA) genes were confirmed by online ARWEN (http://130.235.46.10/ARWEN/) (Laslett and Canback Citation2008). Phylosuite was used for the data processing, including sequences alignment, sequences concatenation (Zhang et al. Citation2020).
The complete mitogenome sequence of C. quinquefasciata was 16,188 in length with A + T content of 84.1%. It consists of 13 protein-coding genes (PCGs), 22 transfer RNAs (tRNAs), 2 ribosomal RNAs (rRNAs), and 2 putative control region (CR). All 13 PCGs were initiated by typical ATN codons (four ATT, five ATG and four ATA). Eleven genes use TAA as terminal stop and two genes stop with T–. All of the 22 tRNA genes, ranging from 58 to 72 bp, have a typical cloverleaf structure except for trnS1, whose dihydrouridine (DHU) arm forms a simple loop. The absence of the DHU arm in trnS1 was found in the mitochondrial genomes existed in most insects (Wolstenholme Citation1992). The control region was 85 and 1032 bp long, with 100% and 81.8% A + T content, respectively. The rrnL and rrnS genes were 1327 and 820 bp, A + T content of them were 84.8% and 86.4%, respectively.
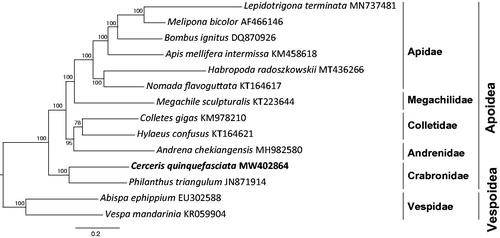
We performed phylogenetic analysis using the 13 PCGs and 2 rRNAs of 12 mitogenome sequences from Apoidea, with two species from Vespidae as outgroups (Abispa ephippium and Vespa mandarinia). Phylogenetic analyses based on 13 PCGs and 2 rRNAs using IQ-TREE (Nguyen et al. Citation2015). The nodes of Bayesian inference phylogeny tree with high support value (). Within superfamily Apoidea, Crabronidae shown a sister relationship with anthophila bees (Apidae, Megachilidae, Colletidae, and Andrenidae) (Branstetter et al. Citation2017). Within Crabronidae, C. quinquefasciata shown a sister relationship with Philanthus triangulum, which both genera belong to the subfamily Philanthinae.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the Accession no. MW402864. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA707660, SRR13906281, and SAMN18219103, respectively.
Additional information
Funding
References
- Bernt M, Donath A, Juhling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Branstetter MG, Danforth BN, Pitts JP, Faircloth BC, Ward PS, Buffington ML, Gates MW, Kula RR, Brady SG. 2017. Phylogenomic insights into the evolution of stinging wasps and the origins of ants and bees. Curr Biol. 27(7):1019–1025.
- Edwards M. 1996. Optimizing habitats for bees in the United Kingdom – a review of recent conservation action. In: A Matheson, SL Buchmann, C O'toole, P Westrich, IH Williams, editors. The conservation of bees. London: Academic Press. p. 35–45.
- Laslett D, Canback B. 2008. ARWEN: a program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics. 24(2):172–175.
- Meng GL, Li YY, Yang CT, Liu SL. 2019. MitoZ: a toolkit for animal mitochondrial genome assembly, annotation and visualization. Nucleic Acids Res. 47(11):e63.
- Nguyen LT, Schmidt HA, Von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Wolstenholme DR. 1992. Genetic novelties in mitochondrial genomes of multicellular animals. Curr Opin Genet Dev. 2(6):918–925.
- Zhang D, Gao F, Jakovlić I, Zou H, Zhang J, Li WX, Wang GT. 2020. PhyloSuite: an integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol Ecol Resour. 20(1) :348–355.