Abstract
Peristrophe japonica (Thunb.) Bremek. is a widely distributed medicinal plant species in China and Japan. In this study, the complete chloroplast genome sequence of P. japonica was assembled and characterized from high-throughput sequencing data. The chloroplast genome is 151,374 bp in length, consisting of a large single-copy (LSC) and a small single-copy (SSC) regions of 83,395 bp and 17,073 bp, respectively, which were separated by a pair of 25,453 bp inverted repeat (IR) regions. The overall GC content of the genome is 38.07%. The genome contains 133 genes, including 88 protein-coding, 37 tRNA, and eight rRNA genes. A phylogenetic tree reconstructed using 23 chloroplast genomes reveals that Peristrophe form a separate group which is a sister of the genus Dicliptera. The work reported here is the first complete chloroplast genome of P. japonica which will provide useful information to the evolutionary studies on the genus of Peristrophe.
Peristrophe japonica (Thunb.) Bremek., with little pink flowers and beautiful slender shape, is a ground covering perennial (Chen et al. Citation2016). P. japonica is widely distributed in China and Japan. It is well known in the national minority of China as herbal medicine in daily life, and is also to treat colds and fever (He et al. Citation2013). Pharmacological analysis shows that P. japonica has anti-bacterial, anti-inflammatory, and cough relief efficacy (Li et al. Citation2010). In addition to medicine, it is successfully used in landscape areas for soil and water conservation. Previous studies on P. japonica mainly focused on the medical effect or the cultivation techniques. However, the chloroplast genome information of P. japonica has not been characterized. In this study, the complete chloroplast genome of P. japonica was determined using high throughput sequencing technology to contribute to the bioinformatics and evolutionary for the phylogenetics of the genus Peristrophe.
The fresh leaves of P. japonica were sampled from Nanjing city, Jiangsu Province, China (31°54′10.368″ N, 118°55′5.12″ E). Specimens were stored in the Medicinal Botanical Garden of China Pharmaceutical University (accession number: CPU-JTSZC, Minjian Qin, [email protected]). Total genomic DNA was extracted with a modified CTAB protocol according to Doyle and Doyle (Citation1987). The whole genome sequencing was conducted by Hefei Biodata Biotechnologies Inc. (Hefei, China) on the Illumina Hiseq platform. The filtered sequences were assembled using the program SPAdes assembler 3.10.0 using the default settings (Bankevich et al. 2012). The annotation was performed with DOGMA (Wyman et al. Citation2004) and BLAST searches.
The chloroplast genome of P. japonica is 151,374 bp in length (GenBank accession no. MW411448), and contains two inverted repeat (IR) regions of 25,453 bp, separated by large single-copy (LSC) and small single-copy (SSC) regions of 83,395 bp and 17,073 bp, respectively. The overall GC content of the P. japonica complete chloroplast genome is 38.07% and the corresponding values in LSC, SSC, and IR regions are 36.1%, 32.1%, and 43.3%, respectively. The complete chloroplast genome was predicted to contain 133 genes, including 88 protein-coding, 37 tRNA, and eight rRNA genes. Nine protein-coding, six tRNA, and four rRNA genes were duplicated in the IR regions. Nineteen genes contained two exons and four genes (clpP, ycf3, and two rps12) contained three exons.
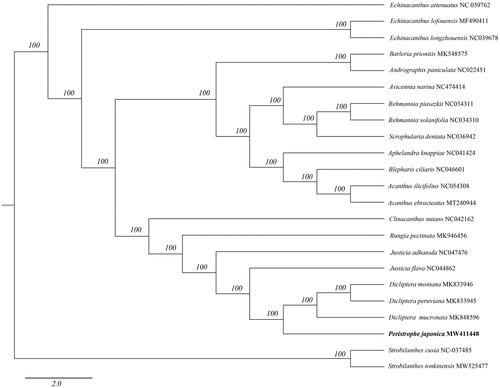
To investigate its taxonomic status, alignment was performed on the 23 complete chloroplast genome sequences which are all from Acanthaceae using MAFFT v7.307 (Katoh and Standley Citation2013), and a maximum-likelihood (ML) tree was constructed with 1000 bootstrap replicates in FastTree v2.1.10 (Price et al. Citation2010) with the GTR + Gamma model. Strobilanthes tonkinensis and S. cusia are as outgroups to construct the phylogenetic tree. As previously published by Deng et al. (Citation2016) Peristrophe form a separate group which is a sister of the genus Dicliptera (). There has no complete chloroplast genome sequence of other species in Peristrophe for reference. Hence, more genome studies of other species in Peristrophe would be necessary for detailed taxonomy research. The complete chloroplast genome sequence of P. japonica will provide a useful resource for the conservation genetics of this species as well as for the phylogenetic studies of the Acanthaceae.
Disclosure statement
The authors report no conflict of interest.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under the accession no. MW411448. The associated BioProject, SRA, and Bio-sample numbers are PRJNA685332, SRS7884003, and SAMN17082919, respectively.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477. doi:10.1089/cmb.2012.0021. 22506599
- Chen JY, Zhu Y, Zhao SW. 2016. Influence of organic mulching on soil environment and growth of Peristrophe japonica. Chin Agric Sci Bull. 32(31):115–122.
- Deng YF, Gao CM, Xia NH, Peng H. 2016. Wuacanthus (Acanthaceae), a new Chinese endemic genus segregated from Justicia (Acanthaceae). Plant Divers. 38(6):312–321.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- He P, Chen L, Zhao C, Ji G. 2013. The research progress of Peristrophe japonica. J Guiyang Coll Tradit Chin Med. 35(1):239–240.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Li Q, Fang SL, Zhao BQ. 2010. The pharmacognostics study of leaves and flowers of Peristrophe japonica. Li Shizhen Med Mater Med Res. 21(12):3154–3155.
- Price MN, Dehal PS, Arkin AP. 2010. FastTree 2 – approximately maximum-likelihood trees for large alignments. PLOS One. 5(3):e9490.
- Wang L, Lu X, Han B, Chen J, Qinlin Lv. 2020. The complete chloroplast genome of Amana baohuaensis (Liliaceae). Mitochondrial DNA Part B. 5(3):3665–3667.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20(17):3252–3255.