Abstract
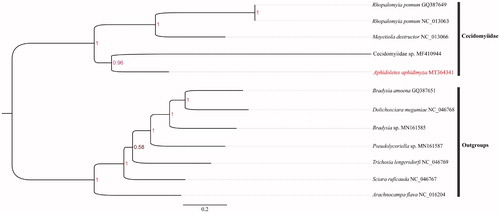
The gall midge of Aphidoletes aphidimyza is ubiquitous, which larva is an effective predator of aphids and entomophagous arthropods used for aphids control. The complete mitogenome of A. aphidimyza was 14,982 bp in length, with 37 typical genes, including 13 protein-coding genes (PCGs), 22 tRNA genes (tRNAs), two rRNA genes (rRNAs). The gene order was consistent with those of known Cecidomyiidae species. All PCGs started with ATN and stopped with TAA. The nucleotide composition was highly A + T biased, accounting for ∼85% of the whole mitogenome. Phylogenetic results show that A. aphidimyza was clustered into the family Cecidomyiidae, which is sister to Cecidomyiidae sp. (MF410944).
The predatory gall midge Aphidoletes aphidimyza (Rondani) (Diptera: Cecidomyiidae) is widely distributed in the world (Harris Citation1973; Yang and Wang Citation1987; Yukawa et al. Citation1998), which is one of the most important entomophagous arthropods used for aphid control. Actually, its larva could prey on over 85 aphid species (Meadow et al. Citation1985; Shang et al. Citation2019; Boulanger et al. Citation2019). In this study, we sequenced and analyzed the mitogenome of Aphidoletes aphidimyza, firstly.
The research materials were collected from Bozhou District, Guizhou, China (106°17′E, 26°57′N) in May 2015. Voucher specimens are deposited in the Institute of Entomology, Guizhou University,China(Maofa Yang, [email protected]) and the laber number is GUGC-IDT-0908521. The total genomic DNA was extracted from 20 instar3rd larvaes of A. aphidimyza. using the DNeasy Blood & Tissue Kit (Cat. No. 69504). The mitogenome of A. aphidimyza was sequenced by Illumina NovaSeq6000 platform with 150 bp paired-end (Berry Genomics, Beijing, China). The reads were assembled and annotated using Generous Prime (v2019.1.3). All tRNA genes were identified by ARWEN v1.2 (Laslett and Canbäck Citation2008). The annotated sequences of mitogenome were submitted to GenBank with accession number MT364341. All protein coding genes (PCGs) were aligned using MAFFT algorithm in the TranslatorX (Abascal et al. Citation2010; Katoh et al. Citation2017), and then poorly aligned results were removed by Gblocks 9.1b (Castresana Citation2000; Abascal et al. Citation2010). Phylogenetic trees were reconstructed based on the 1st and 2nd codon positions of 13 PCGs using the GTR + I + G model determined by MrBayes 3.2.7. on Cipres platform among A. aphidimyza and 11 reference species (Ronquist et al. Citation2012; Meng et al. Citation2019).
The complete mitogenome of A. aphidimyza is 14,982 bp in length with a high A + T content of 85.1%, including 13 PCGs, 22 tRNA genes (tRNAs), two rRNA genes (rRNAs) and one control region. Its gene composition and order were the same as other Cecidomyiidae mitogenomes (Beckenbach and Joy Citation2009). All PCGs used standard started codon ATN and stop codon TAA. The length of 22 tRNA ranged from 37 bp (tRNA-A) to 71 bp (tRNA-G). Genes of 16S rRNA and 12S rRNA are 1,273 bp and 766 bp, respectively.
The phylogenetic tree shows that A. aphidimyza was clustered into clade of Cecidomyiidae species, with sister to Cecidomyiidae sp. (MF410944) (). This work further enriched mitogenome database of the family Cecidomyiidae, with facilitating future studies on taxonomy and molecular systematics.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in GenBank at https: https://www.ncbi.nlm.nih.gov/nuccore/MT364341, Associated BioProject, https://www.ncbi.nlm.nih.gov/bioproject/PRJNA718061, BioSample accession number at https://www.ncbi.nlm.nih.gov/biosample/SAMN18521476 and Sequence Read Archive at https://www.ncbi.nlm.nih.gov/sra/SRR14087947.
Additional information
Funding
References
- Abascal F, Zardoya R, Telford MJ. 2010. TranslatorX: multiple alignment of nucleotide sequence guided by amino acid translations. Nucleic Acids Res. 38:W7–W13.
- Beckenbach AT, Joy JB. 2009. Evolution of the mitochondrial genomes of gall midges (Diptera: Cecidomyiidae): rearrangement and severe truncation of tRNA genes. Genome Biol Evol. 1:278–287.
- Boulanger FX, Jandricic S, Bolckmans K, Wäckers FL, Pekas A. 2019. Optimizing aphid biocontrol with the predator Aphidoletes aphidimyza, based on biology and ecology. Pest Manag Sci. 75:1479–1493.
- Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 17:540–552.
- Harris KM. 1973. Aphidophagous Cecidomyiidae (Diptera): taxonomy, biology and assessments of field populations. Bull Entomol Res. 63(2):305–325.
- Katoh K, Rozewicki J, Yamada KD. 2017. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform. 20(4):1160–1166.
- Laslett D, Canbäck B. 2008. ARWEN: a program to detect tRNA genes in metazoan mitochondrial nucleotide sequence. Bioinform. 24(2):172–175.
- Meadow RH, Kelly WC, Shelton AM. 1985. Evaluation of Aphidoletes aphidimyza (Diptera: Cecidomyiidae) for control of Myzus persicae (Hemiptera: Aphididae) in greenhouse and field experiments in the United States. Entomophaga. 30(4):385–392.
- Meng GL, Li YY, Yang CT, Liu SL. 2019. MitoZ: a toolkit for animal mitochondrial genome assembly, annotation and visualization. Nucleic Acids Res. 47(11):e63.
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biol. 61:539–542.
- Shang SH, Huang CY, Shen XX, Yu XF, Cao Y, Liu MH, Yang MF. 2019. Field control effect of Aphidoletes aphidimyza on tobacco Myzus persicae. Guizhou Agric Sci. 47(6):41–44.
- Yang HF, Wang HZ. 1987. Note on Aphidoletes aphidimyza (Cecidomyiidae) from Xinjiang, China. J Aug First Agr Coll. 4:57–59.
- Yukawa J, Yamaguchi D, Mizota K, Setokuchi O. 1998. Distribution and host range of an aphidophagous species of Cecidomyiidae, Aphidoletes aphidimyza (Diptera) in Japan. Appl Entomol Zool. 33(1):185–193.