Abstract
The complete mitochondrial genome of the Barcheek trevally, Carangoides plagiotaenia Bleeker, 1857 from Beqa lagoon, Fiji Islands, was determined by high-throughput sequencing (HTS). Its mitogenome (16,551 bp) contained the typical 37 genes, including 13 protein-coding genes (PCGs), 2 ribosomal RNAs, 22 tRNAs, along with two non-coding regions: control region (D-loop) and the origin of light-strand replication (OL). In its mitogenome, one unusual start codon (GTG) was identified in COI and incomplete stop codons (TA-/T–) were predicted in seven PCGs including ND2, COII, ATP6, COIII, ND3, ND4 and CytB. The phylogenetic tree revealed that C. plagiotaenia is sister to C. bajad with 88.75% nucleotide identity in 13 PCGs and 90.49% in the complete mitogenomes.
Fish in the family Carangidae, commonly known as trevallies or jacks, are widely distributed from the tropics to the subtropical oceans. According to FishBase (www.fishbase.org), 143 species in 30 genera are currently known in the family. Carangoides plagiotaenia Bleeker, 1857 is distributed throughout the tropical and subtropical waters of the Indian and West Pacific Ocean. This species can be distinguished from its relatives by the fully scaled breast with a black posterior edge preoperculum and a projected lower jaw (Gunn Citation1990). Among 21 species in the genus Carangoides, only four complete mitochondrial genomes are currently reported in the database, including C. bajad, C. armatus, C. malabarius, and C. equula (Zou and Li Citation2016; Song et al. Citation2020) The genetic information of C. plagiotaenia is strongly required for the scientific management of its resources distinctly from its relatives in Fiji. We here report the mitochondrial genome sequence of Caragoides plagiotaenia collected from Fiji using the high-throughput sequencing (HTS) technique.
C. plagiotaenia was caught from Beqa lagoon (18°23′23″S, 178°02′42″E) in Fiji in 2019 and its muscle tissue and DNA are stored at the Marine Biodiversity Institute of Korea (https://www.mabik.re.kr/html/en/, Ha Yeun Song, and [email protected]) under the number MABIK GR00004008. Its COI sequence showed 99.6% sequence identity to Carangoides plagiotaenia (KC970456) confirming the correct identification of the specimen. A library was constructed and read using Truseq® DNA library preparation kit V2 (Illumina, San Diego, CA, USA) and MiSeq system generating 4 million raw reads with 150 bp in length (Illumina, San Diego, CA, USA), respectively. The raw reads were assembled and annotated by Geneious software using the mitogenome of C. armatus (NC_004405) as a template (Kearse et al. Citation2012). The tRNA structures were predicted by tRNAScan-SE (Lowe and Chan Citation2016). The phylogenetic tree was constructed using MEGA X software with a Maximum Likelihood (ML) algorithm and 1,000 bootstrap replicates (Kumar et al. Citation2016). Siniperca scherzeri in the family Sinipercidae was used as an outgroup for the analysis.
The mitochondrial genome of C. plagiotaenia (MT677872) was 16,551 bp in length, which harbors 37 genes (13 protein-coding genes, 2 ribosomal RNAs, and 22 tRNAs). Two non-coding regions, the control region (D-loop) and the origin of light-strand replication (38 bp, OL) were also identified. ND6 and other 8 tRNA genes were encoded on the light (L) strand, while the other 28 genes were located on the heavy (H) strand. Besides COI (GTG), the other twelve protein-coding genes (PCGs) have the canonical start codon (ATG). Typical stop codons (TAG and TAA) were identified in six PCGs, including ND1, ND5, ND6, COI, ND4L and ATP8, whereas incomplete ones (TA-/T–) were also identified in seven PCGs, including ND2, COII, ATP6, COIII, ND3, ND4 and CytB.
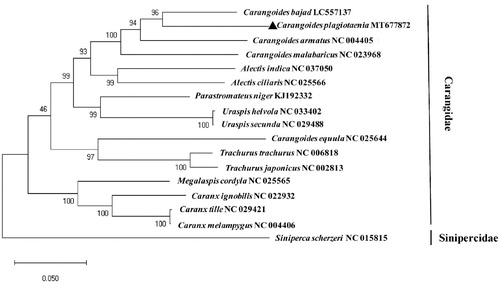
A ML algorithm phylogenetic analysis with 13 PCGs of 16 mitochondrial genomes of the species closely relatied to C. plagiotaenia (). Among those in the Carangidae species, C.plagiotaenia was sister to C. bajad with 88.75% identity, followed by C. armatus (86.99%) and C. malabaricus (86.57%). However, C. equula forms a clade distinct to the cluster of other members in the genus Carangoides suggesting reexamination of its phylogeny in the family Carangidae. More mitogenome sequences should be reported for better understanding of the complicate evolutional relationship in the family Carangidae.
Disclosure statement
The authors report that they have no conflicts of interest. The authors are responsible for the content and writing of the paper.
Data availability statement
The data that support the findings of this study are available in [GenBank database] [Carangoidea plagiotaenia] at https://www.ncbi.nlm.nih.gov/nuccore/MT677872.
Associated accession numbers BioProject: PRJNA670018 and BioSample: SAMN16481768 SRA: SRR12846198 are available at NCBI websites:
https://www.ncbi.nlm.nih.gov/bioproject/PRJNA670018
Additional information
Funding
References
- Gunn JS. 1990. A revision of selected genera of the family Carangidae (Pisces) from Australian waters. Rec Aust Mus, Suppl. 12:1–77.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874.
- Lowe TM, Chan PP. 2016. tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 44:W54–W57.
- Song HY, Jung YH, Choi YJ, Kim B, Nguyen TV, Lee DS. 2020. Complete mitochondrial genome of the orange-spotted trevally, Carangoides bajad (Perciformes, Carangidae) and a comparative analysis with other Carangidae species. Mitochondrial DNA B Resources. 5:3120–3121.
- Zou K, Li M. 2016. Characterization of the mitochondrial genome of the Whitefin trevally Carangoides equula (Perciformes: Carangidae): a novel initiation codon for ATP6 gene. Mitochondrial DNA A DNA Mapp Seq Anal. 27:1779–1780.