Abstract
Octodonta nipae (Maulik 1921) is a dangerous forestry quarantine pest, which mainly harms palms. In the present study, we determined complete mitogenome of O. nipae. This mitogenome was 15,397 bp in length (GenBank Accession no. MW802252), which contained 2 ribosomal RNA genes, 22 transfer RNAs, 13 protein-coding genes (PCGs) and one non-coding AT-rich region with the length of 883 bp. All of the 22 tRNA genes displayed a typical clover-leaf structure, with the exception of tRNAPhe, tRNALeu, tRNAAsn, tRNAPro and tRNAThr. Twelve PCGs were initiated by ATN codons, and NAD1 started with TTG. Ten PCGs used the typical stop codon ‘TAA’ and ‘TGA’, while three PCGs (COX2, COX3, NAD4) used the incomplete stop codons ‘TA’ or ‘T’. Phylogenetic tree demonstrated that O. nipae belongs to the family Chrysomelidae and closer to the superfamily Cassidinae.
Octodonta nipae (Maulik 1921), a dangerous quarantine pest for forestry originating from Malaysia (Fu et al. Citation2020). The adults and larvae of O. nipae gather together to feed on young stems, young shoots and heart leaves of palm plants (Yu et al. Citation2009). Since first discovered in 2001 in China, it has caused serious damage to nursery stock for palm plants, ecological environment, and city afforestation (Fu et al. Citation2020). To date, no mitogenome has been studied for this beetle.
In this paper, individuals of O. nipae were collected from Xixiu Beach Park in Haikou (110°15′36″N, 20°1′48″E), Hainan, China. The specimens were deposited at −20 °C in the herbarium of Post-Entry Quarantine Station for Tropical Plant, Haikou Customs District, PR China (Meng Rui, [email protected]), under the Accession no. IN06091001-0001-0003. Individual hind legs were used to extract DNA. The mitogenome sequence of O. nipae was generated using Illumina HiSeq X TEN Sequencing System and assembled by MitoZ software without parameters (Meng et al. Citation2019).
The complete circular mitogenome of O. nipae had a length of 15,397 bp (Genbank Accession no. MW802252). The nucleotide composition of O. nipae mitogenome was biased toward AT at 74.1%, and the total base composition was 40.0% A, 34.1% T, 9.3% G, and 16.6% C. The circular genome contained two ribosomal RNA genes, 22 transfer RNAs, 13 protein-coding genes (PCGs), and 1 non-coding AT-rich region with the length of 883 bp. The order and orientation of the above mitochondrial genes were typical of other leaf beetle and identical to those of the ancestral insects (Guo et al. Citation2017; Liu et al. Citation2018; Meng et al. Citation2019).
Twelve PCGs started with ATN, except that NAD1 initiated with TTG. 10 PCGs used the typical stop codon ‘TAA’ and ‘TAG’, while three PCGs (COX2, COX3, NAD4) used the incomplete stop codons ‘TA’ or ‘T’. All of the 22 tRNAs have a typical clover-leaf secondary structure, except for tRNAPhe, tRNALeu, tRNAAsn, tRNAPro, and tRNAThr whose TΨC arm formed a simple loop. All tRNAs had normal lengths, which varied from 61 to 70 bp. The 16S rRNA was 1338 bp long with an AT content of 78.6%, while the 12S rRNA was 792 bp long with an AT content of 78.0%. The non-coding region (putative control region) was 883 bp that located between 12S rRNA and tRNAIle.
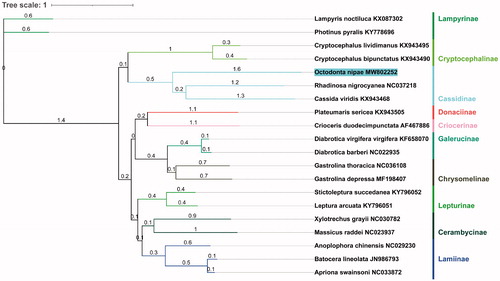
The concatenated datasets of the 13 PCGs from mitogenome of 18 Chrysomeloidea species from NCBI were adopted to build phylogenetic tree. Lampyris noctiluca and Photinus pyralisd that from Elateroidea were used as outgroups. The analyses were performed with Bayesian inference in Phylosuite (Ronquist et al. Citation2012; Zhang et al. Citation2020). The phylogenetic tree () demonstrated O. nipae belongs to the family Chrysomelidae and closer to the superfamily Cassidinae. In conclusion, the results of this study can provide essential and important DNA molecular data for further phylogenetic and evolutionary analysis of Chrysomeloidea.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession No. MW802252. The associated BioProject, Bio-Sample and SRA numbers are PRJNA723914, SAMN18837207 and SRR14306651, respectively.
Additional information
Funding
References
- Fu L, Tang BZ, Hou YM. 2020. Progress in research on Octodonta nipae, an invasive pest of palms. J Environ Entomol. 42(4):829–837.
- Guo QY, Xu JS, Dai XH, Liao CQ, Long CP. 2017. Complete mitochondrial genome of a leaf-mining beetle, Rhadinosa nigrocyanea (Coleoptera: Chrysomelidae) with phylogenetic consideration. Mitochondrial DNA Part B. 2(2):446–448.
- Liu P, Guo QY, Xu JS, Liao CQ, Dai XH. 2018. Complete mitochondrial genome of a leaf beetle, Callispa bowringi (Coleoptera: Chrysomelidae). Mitochondrial DNA Part B. 3(1):213–214.
- Meng GL, Li YY, Yang CT, Liu SL. 2019. MitoZ: a toolkit for animal mitochondrial genome assembly, annotation and visualization. Nucleic Acids Res. 47(11):e63–e363.
- Meng R, Ao S, Xu W, Lv BQ, Cai B. 2019. Complete mitochondrial genome of coconut hispine beetle Brontispa longissima (Coleoptera: Chrysomelidae: Cassidinae). Mitochondrial DNA Part B. 1(5):1126–1127.
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542.
- Yu FY, Qin WQ, Ma ZL, Li ZX, Huang SC, Han CW. 2009. Effects of host plants on development and fecundity of Octodonta nipae. Plant Prot. 35(2):72–74.
- Zhang D, Gao FL, Li WX, Jakovlić I, Zou H, Zhang J, Wang GT. 2020. PhyloSuite: an integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol Ecol Resour. 20(1):348–355.