Abstract
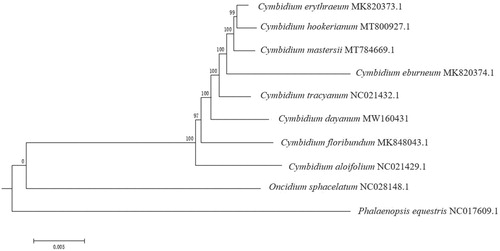
Cymbidium dayanum, a wild orchid species in the Orchid family (Orchidaceae), is considered highly valuable because of its long flowering period and beautiful plant shape. We sequenced the complete chloroplast genome of C. dayanum using the Illumina Hiseq platform (Illumina, San Diego, CA). The size of the C. dayanum chloroplast genome is 155,408 bp, with an average GC content of 36.76%. This chloroplast genome has containing a large single-copy (LSC) region of 84,189 bp, a small single-copy (SSC) region of 17,991 bp, and two inverted (IRa and IRb) repeat regions of two 26,614 bp. A total of 118 unique genes were annotated, including 76 protein-coding genes, 38 tRNA genes, and 4 rRNA genes. A maximum-likelihood phylogenetic tree indicated that C. dayanum is closely related to C. tracyanum in the genus Cymbidium based on 9 whole chloroplast genome sequences.
Cymbidium dayanum is an epiphytic orchid, it mainly grows on trees in open forests or cliffs along streamsides, with altitude ranging between 300 and 1600 m. The flowers have white or cream-yellow sepals and petals, with a maroon stripe in the center (Liu et al. Citation2006). The flowering period spans for approximately 4 months, from fall to winter, with the inflorescences often blooming asynchronously on each plant and individual flowers commonly lasting for a month (Matsuda and Sugiura Citation2019). Because of its long flowering period and beautiful plant shape, C. dayanum is considered a very valuable wild orchid. Cymbidium dayanum has great utilities as a breeding parent of orchids because of its arching and pendulous inflorescence, its utilization as a parent for hybridization of Cymbidium hybrid, and successful intergeneric hybridization of C. dayanum has been previously recorded (The International Orchid Register: http://apps.rhs.org.uk/horticulturaldatabase/orchidregister). Cymbidium dayanum is widely distributed, from Northern India to Japan, South-Eastern Asia and Sumatra (Du and Cribb 2007). In recent years, due to the gradual destruction of ecological environment and excessive collection, the wild population of C. dayanum has been drastically reduced (Luo et al. Citation2008). Therefore, resource conservation and genetic breeding research of C. dayanum are in urgent needs, since information about the genome of C. dayanum is scarce. In this study, the completed chloroplast genome of C. dayanum has been reported for the first time based on illumina pair-end sequencing data, which is useful for genetic utilization of germplasm resources of the family Cymbidium in the future.
The fresh samples of C. dayanum were collected from Xinyi county of Guangdong province, China(110°57′04″E, 22°21′10″N), and the voucher specimen of C. dayanum was stored at the South China Botanical Garden, Chinese Academy of Sciences (Accession no. 14916, e-mail: [email protected]). Total genomic DNA was extracted from fresh leaf tissue using the modified CTAB method (Doyle and Doyle Citation1987), and the isolated DNA was manufactured to average 350 bp paired-end libraries using NEBNext Ultra DNA Library Prep Kit (Illumina, Beijing, China). The constructed libraries were sequenced using NovaSeq platform provided by Benagen Tech Solutions (Wuhan, China), and the collected raw sequences data were quality controlled and removed by the FastQC (Cock et al. Citation2010). After filtering, approximately 2.2 GB paired-end clean reads were assembled with the program SPAdes (version: 3.13.0) using the Cymbidium aloifolium chloroplast genome sequence as reference (Bankevich et al. Citation2012). Annotations were performed using the online program CPGAVAS2 (Shi et al. Citation2019). The C. dayanum chloroplast genome sequence was submitted to GenBank (Accession no. MW160431).
The complete chloroplast genome sequence of C. dayanum was 155,408 bp in length, with a large single-copy (LSC) region of 84,189 bp, a small single-copy (SSC) region of 17,991 bp, and a pair of inverted repeats (IR) regions of 26,614 bp. The overall GC content was 36.76%, whereas the corresponding values of the LSC, SSC, and IR regions were 34.27%, 29.36%, and 43.21%, respectively. A total of 136 gene species were annotated, including 76 protein-coding genes (PCG), 38 transfer RNA (tRNA), and 4 ribosomal RNA (rRNA) gene species. There were 18 genes duplicated in the IR regions, including 6 PCG genes, 8 tRNA genes, and 4 rRNA genes, and the remaining 118 genes were unique genes. Twelve genes contain one intron, while three genes have two introns.
To confirm the phylogenetic status of C. dayanum, a molecular phylogenetic tree was constructed using the maximum-likelihood (ML) methods, based on the complete chloroplast genomes of seven Cymbidium species and two Orchidaceae species as outgroups downloaded from GenBank. ClustalW was applied for multisequence alignment, and the ML analysis was performed using the MEGA6 under the best model of Tamura-Nei model and bootstrapped with 1000 replicates (Vilella et al. Citation2009). Results showed that ten Orchidaceae species were all clustered together (). The phylogenetic tree indicated that C. dayanum is closely related to C. tracyanum supported with 100 bootstrap support values. The characterized chloroplast genome sequence of C. dayanum will be helpful for further study on the phylogenetic study, species identification, and genetic engineering.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The data that support the findings of this study are openly available in NCBI database at https://www.ncbi.nlm.nih.gov. GenBank Accession no. MW160431, BioProject Accession no. PRJNA718379.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Cock PJ, Fields CJ, Goto N, Heuer ML, Rice PM. 2010. The Sanger FASTQ file format for sequences with quality scores, and the Solexa/Illumina FASTQ variants. Nucleic Acids Res. 38(6):1767.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure from small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Du P, Cribb D. 1988. The genus Cymbidium. London: Christopher Helm.
- Liu ZJ, Chen SC, Ru ZZ. 2006. The genus Cymbidium in China. Beijing: Science Publishing. p. 57–58.
- Luo YH, Leng QY, Mo R, Chen YY. 2008. Nonsymbiotic germination and low-temperature in vitro conservation of Cymbidium dayanum. J Anhui Agric Sci. 36(19):67–69.
- Matsuda Y, Sugiura N. 2019. Specialized pollination by honeybees in Cymbidium dayanum, a fall-winter flowering orchid. Plant Species Biol. 34(1):19–26.
- Shi LC, Chen HM, Jiang M, Wang LQ, Wu X, Huang LF, Liu C. 2019. CPGAVAS2, an integrated plastome sequence annotator and analyzer. Nucleic Acids Res. 47(W1):W65–W73.
- Vilella AJ, Severin J, Ureta-Vidal A, Heng L, Durbin R, Birney E. 2009. EnsemblCompara GeneTrees: complete, duplication-aware phylogenetic trees in vertebrates. Genome Res. 19(2):327–335.