Abstract
The Yellow-legged Buttonquail Turnix tanki is a species of the genus Turnix, which belongs to the order Charadriiformes. It is distributed across almost all of China. The International Union for Conservation of Nature has assessed the bird’s conservation status as ‘Least Concern (LC).’ We sequenced the complete mitogenome of T. tanki and examined its phylogenetic relationship with other charadriiformes species. The mitochondrial DNA is packaged in a compact 17,620 base pair circular molecule with A + T content of 57.90%. It contains 37 typical mitochondrial genes, including 13 protein-coding genes, two rRNAs and 22 tRNAs, and two non-coding regions. We reconstructed a phylogenetic tree based on mitogenome sequences of five Turnicidae species and one outgroup. Phylogenetic analysis indicated that T. tanki is a sister to T. suscitator.
The Yellow-legged Buttonquail Turnix tanki is a species of the genus Turnix, which belongs to the order Charadriiformes. It is reported to have a very large and stable range over 20,000 km2, which places it under the threshold for vulnerable status. It is listed as the Least Concern (LC) species by International Union for Conservation of Nature and Natural Resources (IUCN, Citation2016), supported by the estimated population, which is not believed to approach the thresholds vulnerable status (<10,000 mature individuals with a continuing decline estimated to be >10% in 10 years or three generations (BirdLife International Citation2020)). It has also been listed as a LC species on the Red List of China’s Vertebrates (Jiang et al. Citation2016). They often build their nests in the grass on the ground or in wheat and soybean fields, usually using the shallow pit to build the nest, and covered by the surrounding crops or weeds. The Yellow-legged Buttonquail looks like a quail, but it is smaller. Its back, shoulders, waist, and tail are grayish brown with small black and brown spots. The tail is short and the central tail feather is not elongated. The Yellow-legged Buttonquail is not good at singing, but it runs fast on the ground. It usually runs away from danger rather than taking to the air. It mainly feeds on plant buds, berries, grass seeds, grains, and insects and other small invertebrates (Zhao, Citation2001). Here, we report the complete mitogenome of T. tanki (GenBank number: MW307919) and examined its phylogenetic relationship with other Charadriiformes species whose mtDNA data are available.
The specimen (NO. GZUNZ20201129003; contact person: Xue Gou; email: [email protected]) was collected from Huaxi in the Guiyang on 21 November 2019 (E106°40′19.60″, N26°25′58.42″), and was stored in zoological specimens room of Research Center for Biodiversity and Nature Conservation of Guizhou University. Unilateral legs were taken, exoskeleton was removed carefully, and then muscle tissue was gathered for mtDNA isolation. The mitochondrial genome of Yellow-legged Buttonquail was extracted by the Mitochondrial Extraction Kit (Beijing Aidlab Biotechnologies Co., Ltd.). The mitochondrial genomes of T. velox (MN356355.1) were used to design primers for polymerase chain reaction (PCR) and used as a template for gene annotation.
The complete mtDNA sequence of the Yellow-legged Buttonquail is 17,620 bp in length. Its overall base composition is A, 31.40%; C, 29.80%; G, 12.30% and T, 26.50%. The A + T content is 57.90%. It has a typical circular mitochondrial genome containing 13 protein-coding genes, 22 transfer RNAs, two ribosomal RNAs, and two non-coding A + T-rich regions, which is typical in birds (Boore Citation1999). The order and orientation are identical to the standard avian gene order (Gibb et al. Citation2007). Of the 13 protein-coding genes, 11 utilize the standard mitochondrial start codon ATG. However COI uses GTG and ND3 uses ATT as the initiation codon. TAA is the most frequent stop codon, although COIII, ND3, and ND4 end with the single nucleotide T; COI, ND1, and ND6 end with AGG; and ND2 ends with TAG.
The 12S rRNA is 969 bp in length, and the 16S rRNA is 1590 bp in length, which are located between tRNA-Phe (gaa) and tRNA-Leu (taa), and separated by tRNA-Val (tac). All tRNAs have the classic cloverleaf secondary structure, as observed in other bird mitogenomes (Bernt et al. Citation2012). Most of the mitochondrial genes are encoded on the heavy strand (H-strand), but ND6 and eight tRNA genes are instead encoded on the light strand (L-strand) ().
Table 1. Organization of the complete mitochondrial genome of Yellow-legged Buttonquail Turnix tanki.
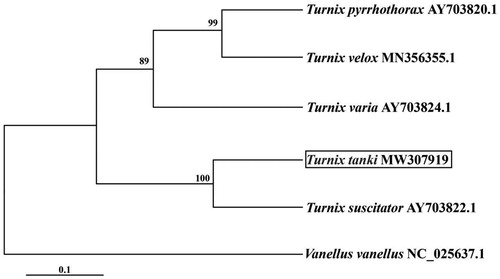
We used MEGA6 to construct a phylogenetic tree using the maximum-likelihood method () based on the mitogenome sequences of T. tanki from the other five Turnicidae species and one outgroup according to a previous study. The results showed T. tanki to be a sister to T. suscitator ().
Figure 1. Maximum likelihood tree based on mitogenome sequences of five Turnicidae species and one outgroup. Bootstrap values were presented under the branch, and the posteriori probability values are omitted because all of them are 100%.
This study is the first to report and analyze the complete mitochondrial genome of Yellow-legged Buttonquail, T. tanki. The complete mitogenome of T. tanki may facilitate conservation of this non-flagship species and provide fundamental genetic data for research into the evolution of Turnicidae.
Acknowledgements
We would like to express our gratitude to editors and the anonymous reviewers for their critical comments on the manuscript.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
Mitogenome data supporting this study are openly available in GenBank at: https://www.ncbi.nlm.nih.gov/nuccore/MW307919. Associated BioProject accession numbers are https://www.ncbi.nlm.nih.gov/bioproject/PRJNA724019.
Additional information
Funding
References
- Bernt M, Braband A, Schierwater B, Stadler PF. 2012. Genetic aspects of mitochondrial genome evolution. Mol Phylogenet Evol. 69:328–338.
- BirdLife International. 2020. Species factsheet: Turdus dissimilis. [accessed 2020 Dec 20]. http://www.birdlife.org
- Boore JL. 1999. Survey and summary animal mitochondrial genomes. Nucleic Acids Res. 27:1767–1780.
- Gibb GC, Kardailsky O, Kimball RT, Braun EL, Penny D. 2007. Mitochondrial genomes and avian phylogeny: complex characters and resolvability without explosive radiations. Mol Biol Evol. 24:269–280.
- IUCN 2016. The IUCN Red List of Threatened Species 2016: e.T22708759A94175329..
- Jiang ZG, Jiang JP, Wang YZ, Zhang E, Zhang YY, Li LL, Xie F, Cai B, Cao L, Zheng GM, Dong L, Zhang ZW, Ding P, Luo ZH, Ding CQ, Ma ZJ, et al. 2016. Red list of China’s vertebrates. Biodivers Conserv. 24:5.
- Zhao ZJ. 2001. The Avifauna of China. Jilin: Jilin Science and Technology Press.
