Abstract
Agave sisalana is one of the Agave cultivars for sisal fiber production around the tropical areas of the world. In the present study, we successfully sequenced and assembled its chloroplast genome. The full size of the genome is 157,268 bp with a GC content at 37.85%. The genome is constructed with a large single copy region (LSC, 85,894 bp), a pair of inverted repeat regions (IR, 26,573 bp) and a small single copy region (SSC, 18,228 bp). Besides, 86 protein-coding genes, 38 tRNAs and 8 rRNAs are annotated on the chloroplast genome. Phylogenetic result reveals that A. sisalana is closely related with A. americana and A. H11648.
Agave plants has been widely cultivated for the production of food, beverage, fiber and medicine around the world (Huang et al. Citation2019). Among the 166 cultivated Agave species, Agave sisalana is commonly used for sisal fiber production (Gil-Vega et al. Citation2006; Huang et al. Citation2018). As an important kind of natural fibers, sisal has several properties of high strength, tough texture and friction resistance, which has been widely used as high strength industrial materials for automotive, navigation, papermaking, etc. (Li et al. Citation2000). Agave plants are the native species in the arid and semi-arid regions in Mexico, which leads to their extreme ecological adaptation (Borland et al. Citation2009). The stability of chloroplast is susceptible to environmental stresses. Several studies have been reported to reveal the mechanism of heat and drought tolerance in Agave plants (Luján et al. Citation2009; Sarwar et al. Citation2019). However, few reports are related to other stresses, such as cold and lead stresses, even if they are harmful to plant chloroplast (Zhou et al. Citation2017; Trentmann et al. Citation2020). Besides, the systematic position and phylogenetic relationship of A. sisalana are still ambiguous at chloroplast (cp) genome level. We hence sequenced the cp genome of Agave sisalana, which could benefit future studies related to Agave chloroplast.
A. sisalana leaves were collected from the germplasm garden (22.90°N, 108.33°E) of Guangxi Subtropical Crops Research Institute, Nanning, China. The specimen was stored in Herbarium of Environment and Plant Protection Institute, Chinese Academy of Tropical Agricultural Sciences (EPPI-jm2020011). Total DNA sample was isolated by the modified CTAB method and then sent to Biozeron Biotech (Shanghai, China) for sequencing by Illumina HiSeq 2500 platform, which generated 6.0 Gb raw data (Doyle and Doyle Citation1987). The raw data was deposited to SRA under the accession number of PRJNA705409. The NOVOPlasty and GapCloser software were used for cp genome assembly and gap filling, respectively (Luo et al. Citation2012; Dierckxsens et al. Citation2017). We further selected the DOGMA software for genome annotation and the Geneiousv11.0.3 software for correction (Wyman et al. Citation2004; Kearse et al. Citation2012). The full cp genome sequence of A. sisalana was uploaded to GenBank under an accession number of MW540497.
The total length of A. sisalana cp genome is 157,268 with a GC content at 37.85%. The genome is constructed with a large single copy region (LSC, 85,894 bp), a pair of inverted repeat regions (IR, 26,573 bp) and a small single copy region (SSC, 18,228 bp). Besides, 86 protein-coding genes, 38 tRNAs and 8 rRNAs are annotated on the cp genome.
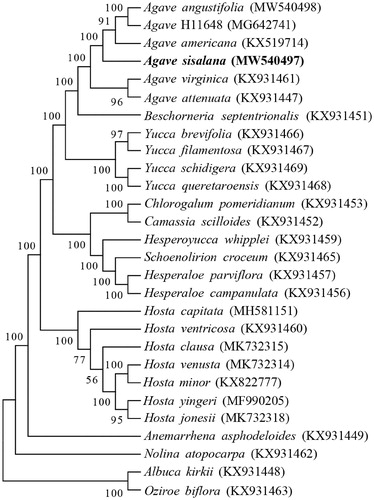
The cp genome sequences of 27 species were selected in phylogenetic analysis. Among these, 24 species were from Agavoideae and 3 other species were selected as outgroup, including Albuca kirkii, Nolina atopocarpa and Oziroe biflora (McKain et al. Citation2016; Lee et al. Citation2019; Jin et al. Citation2020). The alignment of the long sequences was carried out by the MAFFT software (Katoh and Standley Citation2013). The phylogenetic tree was constructed by the Maximum Likelihood method with 1000 bootstrap replicates in MEGA7 software (Kumar et al. Citation2016). The result revealed that A. sisalana is closely related with A. americana, A. angustifolia and A. H11648 (). A. H11648 is an interspecific hybrid from A. angustifolia and A. amaniensis, which is also a widely cultivated Agave species for fiber production (Huang et al. Citation2019). This study would contribute to reveal the mechanism of stress tolerance related to chloroplast in A. sisalana.
Disclosure statement
The authors declare that they have no competing interests.
Data availability statement
The data that support the findings of this study are fully available in SRA (https://www.ncbi.nlm.nih.gov/sra/?term=PRJNA705409) and GenBank (https://www.ncbi.nlm.nih.gov/nuccore/MW540497).
Additional information
Funding
References
- Borland AM, Griffiths H, Hartwell J, Smith JA. 2009. Exploiting the potential of plants with crassulacean acid metabolism for bioenergy production on marginal lands. J Exp Bot. 60:2879–2896.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45:e18.
- Doyle JJ, Doyle JL. 1987. A Rapid DNA isolation procedure from small quantities of fresh leaf tissues. Phytochem Bull. 19:11–15.
- Gil-Vega K, Diaz C, Nava-Cedillo A, Simpson J. 2006. AFLP analysis of Agave tequilana varieties. Plant Sci. 170(4):904–909.
- Huang X, Wang B, Xi J, Zhang Y, He C, Zheng J, Gao J, Chen H, Zhang S, Wu W, et al. 2018. Transcriptome comparison reveals distinct selection patterns in domesticated and wild Agave species, the important CAM plants. Int J Genom. 2018:5716518.
- Huang X, Xiao M, Xi J, He C, Zheng J, Chen H, Gao J, Zhang S, Wu W, Liang Y, et al. 2019. De Novo transcriptome assembly of agave H11648 by Illumina sequencing and identification of cellulose synthase genes in agave species. Genes. 10(2):103.
- Jin G, Huang X, Chen T, Qin X, Xi J, Yi K. 2020. The complete chloroplast genome of agave hybrid 11648. Mitochondrial DNA Part B. 5:2345–2346.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. GeneiousBasic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874.
- Lee SR, Kim K, Lee BY, Lim CE. 2019. Complete chloroplast genomes of all six Hosta species occurring in Korea: molecular structures, comparative, and phylogenetic analyses. BMC Genomics. 20:833.
- Li Y, Mai YW, Ye L. 2000. Sisal fibre and its composites: a review of recent developments. Compos Sci Technol. 60(11):2037–2055.
- Luján R, Lledías F, Martínez LM, Barreto R, Cassab GI, Nieto-Sotelo J. 2009. Small heat-shock proteins and leaf cooling capacity account for the unusual heat tolerance of the central spike leaves in Agave tequilana var. Weber. Plant Cell Environ. 32:1791–1803.
- Luo R, Liu B, Xie Y, Li Z, Huang W, Yuan J, He G, Chen Y, Pan Q, Liu Y, et al. 2012. Soapdenovo2: an empirically improved memory-efficient short-read de novo assembler. GigaScience. 1:1.
- McKain MR, McNeal JR, Kellar PR, Eguiarte LE, Pires JC, Leebens-Mack J. 2016. Timing of rapid diversification and convergent origins of active pollination within Agavoideae (Asparagaceae). Am J Bot. 103:1717–1729.
- Sarwar MB, Ahmad Z, Rashid B, Hassan S, Gregersen PL, Leyva MO, Nagy I, Asp T, Husnain T. 2019. De novo assembly of Agave sisalana transcriptome in response to drought stress provides insight into the tolerance mechanisms. Sci Rep. 9:396.
- Trentmann O, Mühlhaus T, Zimmer D, Sommer F, Schroda M, Haferkamp I, Keller I, Pommerrenig B, Neuhaus HE. 2020. Identification of chloroplast envelope proteins with critical importance for cold acclimation. Plant Physiol. 182:1239–1255.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with dogma. Bioinformatics. 20:3252–3255.
- Zhou J, Jiang Z, Ma J, Yang L, Wei Y. 2017. The effects of lead stress on photosynthetic function and chloroplast ultrastructure of Robinia pseudoacacia seedlings. Environ Sci Pollut Res Int. 24:10718–10726.