Abstract
Prunus sargentii is an ornamental flowering cherry species, spread in Japan, Korea, Russia, and Northeast China. Little information is available regarding its genomic, with limited phylogenetic relationship study performed on P. sargentii until now. In this research, we reported the complete plastid genome of P. sargentii. The complete chloroplast of this species is 158,138 bp in length, including a pair of invert repeat regions (IR) (26,463bp) that is divided by a large single-copy region (LSC) (85,959bp) and a small single-copy region (SSC) (19,253bp). The plastid genome contained a total of 128 genes, including 84 coding genes, eight rRNA genes, and 36 tRNA genes. Phylogenetic analysis indicates that P. sargentii has a closer relationship with P. kumanoensis.
Keywords:
Prunus L. subg. Cerasus (Mill.) A. Gray contains approximately 150 species that mainly occupy temperate and subtropical regions of the northern hemisphere (Yu et al. Citation1986). Since subg. Cerasus provides various edible cherries and ornamentals of economic value, these taxa have great potential for development and application (Li et al. Citation2019). Prunus sargentii Rehder (Schwerin and Beissner Citation1908, p. 159), commonly called Sargent cherry or North Japanese hill cherry, is a kind of graceful ornamental flowering cherry trees. This species is mainly distributed in Japan, Korea, Russia, and Northeast China. Meanwhile, the genetic relationship of P. sargentii relative to other subg. Cerasus is poorly understood. Therefore, we sequenced the whole chloroplast genome of P. sargentii to elucidate its phylogenetic relationship with other subg. Cerasus species.
The plant material was obtained from Dongling, Liaoning province, China (41°83′65″N 123°58′34″E, altitude 90 m). A specimen was deposited at Nanjing Forestry University (https://shengwu.njfu.edu.cn/; collector: Meng Li, [email protected]; voucher number: NF: 161098816). Total DNA was extracted from fresh leaves with a modified CTAB protocol. The whole-genome sequencing was conducted by Nanjing Genepioneer Biotechnologies Inc. (Nanjing, China) on the Illumina Hiseq 2500 platform (Illumina, San Diego, CA). A total of 3.54 Gb clean PE reads (Phred scores >20) were assembled using the program GetOrganelle version 1.7.2 (Jin et al. Citation2020). The plastome was annotated by the web application GeSeq (https://chlorobox.mpimp-golm.mpg.de/geseq.html) (Tillich et al. Citation2017).
The complete circular plastid genome of P. sargentii (GenBank accession No. MW392082) was 158,138 bp in length. Consisting of four regions; large single-copy region (LSC) of 85,959 bp, small single-copy region (SSC) of 19,253 bp, and a pair of inverted repeat regions (IRA and IRB) of 26,463 bp each. The overall GC contents of the plastid genome were 36.7%; LSC (34.6%), SSC (30.3%), and IR (42.5%). The genome contained a total of 128 genes, including 84 coding genes, eight rRNA genes, and 36 tRNA genes.
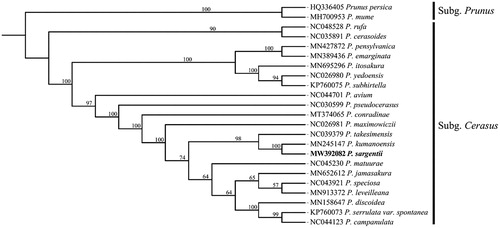
Phylogenetic analysis including P. sargentii, 20 other subg. Cerasus species and two outgroups of subg. Prunus were performed using complete plastid genomes. Sequences were aligned by MAFFT version 7.467 (Katoh et al. Citation2005) and visually checked and adjusted in Bioedit. Maximum-likelihood (ML) analysis was conducted in IQ-TREE version 2.1.1 (Vergara et al. Citation2015). The result was well-resolved and revealed that P. sargentii was belonged to subg. Cerasus and most closely related to P. kumanoensis (). In summary, the complete plastid genome of P. sargentii will provide useful genetic information for increasing the richness of subg. Cerasus, as well as assisting in phylogenetic and evolutionary studies of subg. Cerasus.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The plastid genome in this study is available in the NCBI GenBank (https://www.ncbi.nlm.nih.gov/genbank/) with an accession number MW392082. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA686839, SRR13279544, and SAMN17126435, respectively.
Additional information
Funding
References
- Jin JJ, Yu WB, Yang JB, Song Y, Depamphilis CW, Yi TS, Li DZ. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21:241.
- Katoh K, Kuma KI, Toh H, Miyata T. 2005. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 33(2):511–518.
- Li M, Song YF, Zhu SX, Zhu H, Yi XG, Chen ZC, Li MZ, Wang XR. 2019. The complete plastid genome of cherry plants Prunus discoidea (Rosaceae) and its phylogenetic implication. Mitochondrial DNA Part B. 4:3640–3641.
- Schwerin GV, Beissner L. 1908. Mitteilungen der Deutschen Dendrologischen Gesellschaft. Vol. 17. Berlin (Germany); p. 159.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45:W6–W11.
- Vergara D, White K, Keepers K, Kane N. 2015. The complete chloroplast genomes of Cannabis sativa and Humulus lupulus. Mitochondrial DNA. 27:1–2.
- Yu TT, Lu LT, Ku TC, Li CL, Chen SX. 1986. Rosaceae (3) Prunoideae. In: Yu TT, editor. Flora Reipublicae Popularis Sinicae, Tomus. Beijing, China: Science Press; p. 38.