Abstract
The first complete mitochondrial genome of Euroleon coreanus (Okamoto, 1926) was 15,797 bp in length, and contained 13 protein-coding genes, 22 transfer RNAs, two ribosomal RNAs, and the control region. Compared to the classic insect mitochondrial genome, E. coreanus showed a gene rearrangement of ND2-C-W-Y-COX1. The overall AT content of the mitochondrial genome was 75.5%. The monophyly of Ascalaphidae, Myrmeleontidae, Nemopteridae, Nymphidae, and Psychopsidae was supported in both BI and ML trees. And E. coreanus was a sister clade to the clade of genus Myrmeleon.
Neuroptera is an early-diverging lineage of holometabolous insects including about 6,000 species in 19 families (Song et al. Citation2019; Oswald Citation2020). Myrmeleontidae is one of the most species-rich families within Neuroptera (Machado et al. Citation2019; Machado and Oswald Citation2020; Zheng and Liu Citation2021) The phylogenetic relationship between Myrmeleontidae and Ascalaphidae was controversial in past studies that were based on morphological and molecular data (Aspöck et al. Citation2001; Winterton et al. Citation2010; Yan et al. Citation2014; Wang et al. Citation2017; Zhang and Yang Citation2017; Song et al. Citation2018; Winterton et al. Citation2018). Euroleon coreanus (Okamoto, 1926) is currently assigned to the subfamily Myrmeleontinae of Myrmeleontidae, which is widely distributed in Korea, Mongolia, and North China (Liu et al. Citation2016). Besides, the larvae can be treated as traditional Chinese medicine (Liu et al. Citation2016). In this study, we sequenced the complete mitochondrial genome of E. coreanus to enrich the molecular data of Myrmeleontidae, and further discuss the phylogenetic status of Myrmeleontidae.
The sample of E. coreanus was collected from Yitong Manchu Autonomous County (N 43.200°, E 125.170°), Jilin Province, China, then identified by Dr. JY Zhang. The specimen under the voucher number: JLYT20190826-1 was preserved in a −40 °C freezer and deposited at the Animal Specimen Museum, College of Life Sciences and Chemistry, Zhejiang Normal University, China (sky.zjnu.edu.cn, DN Yu, [email protected]). Total DNA was extracted from muscle tissue of forelegs using an Ezup Column Animal Genomic DNA Purification Kit (Sangon Biotech Company, Shanghai, China) and stored in the Zhang laboratory. We used usual primers to amplify several partial fragments (Simon et al. Citation2006) and designed species-specific primers based on previously obtained sequences using Primer Premier 5.0 (Lalitha Citation2000). All PCR products were then sequenced using the primer-walking method by Sangon Biotech Company (Shanghai, China). The tRNA genes were identified using the MITOS web server (http://mitos.bioinf.uni-leipzig.de/index.py) (Bernt et al. Citation2013). Thirteen PCGs and two rRNA genes (12S and 16S rRNA) were identified by aligning with homologous gene sequences of other Myrmeleontidae species using Mega 7.0 (Kumar et al. Citation2016). In addition, we translated the nucleotide sequences of the 13 protein-coding genes (PCGs) into amino acids based on the invertebrate genetic codes. The E. coreanus mitochondrial genome was deposited in GenBank with an accession number MW879174.
The complete mitochondrial genome of E. coreanus was 15,797 bp in length, including 13 protein-coding genes (PCGs), 22 transfer RNAs, two ribosomal RNAs, and a control region (982 bp). The nucleotide composition of E. coreanus was as follows: A = 39.2%, T = 36.3%, C = 14.6%, G = 9.9%. The overall AT content of the mitochondrial genome was 75.5%. Compared to the classic insect mitochondrial gene sequence, the trnW gene had moved into a position between trnC and trnY, which gave a gene order of ND2-C-W-Y-COX1. However, this gene arrangement is common in the mitochondrial genomes of Myrmeleontidae (Song et al. Citation2019). For the 13 PCGs, almost all used ATN as the initiation codon, excluding COX1 that used ACG. COX1, COX2 and ND5 used stop codons that were incomplete (T-) whereas the remaining 10 protein-coding genes ended with typical stop codons (TAA or TAG). Four protein-coding genes (ND1, ND4, ND4L, ND5), eight tRNA genes (tRNAGln, tRNACys, tRNATyr, tRNAPhe, tRNAHis, tRNAPro, tRNALeu1, tRNAVal) and two rRNA genes (16S rRNA, 12S rRNA) were located on the light strand (L strand), the remaining 23 genes were coded on the heavy strand (H strand). Overall, nine gene overlaps and 11 gene-spacing regions between adjacent genes were found in the mitochondrial genome of E. coreanus, ranging from 1 to 7 bp and 1 to 69 bp in length, respectively.
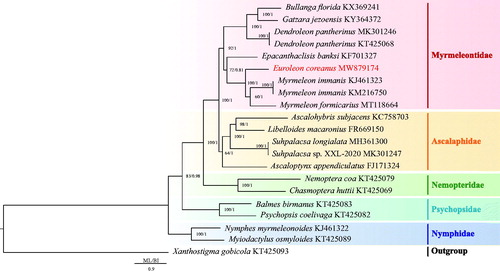
Based on the 13 PCGs, the phylogenetic relationship was constructed using two methods: Bayesian inference (BI) using MrBayes 3.1.2 (Huelsenbeck and Ronquist Citation2001) and Maximum-likelihood (ML) using RAxML 8.2.0 (Stamatakis Citation2014). The mitochondrial genomes of 21 Neuroptera species including E. coreanus and 20 species downloaded from GenBank were used to investigate the phylogenetic relationships within Neuroptera (Beckenbach and Stewart Citation2009; Negrisolo et al. Citation2011; Cheng et al. Citation2014; Yan et al. Citation2014; Cheng et al. Citation2015; Lan et al. Citation2016; Zhang and Wang Citation2016; Zhang and Yang Citation2017; Gao et al. Citation2018; Song et al. Citation2019; Wu et al. Citation2020), with Xanthostigma gobicola chosen as the outgroup. The results of the two phylogenetic analyses (BI and ML) using the 13 protein-coding genes yielded the same topological structure (). The monophyly of Ascalaphidae, Myrmeleontidae, Nemopteridae, Nymphidae, and Psychopsidae was supported in both BI and ML trees. Nymphidae was the basal clade of Neuroptera and then the phylogenetic relationship of (Nymphidae + (Psychopsidae + (Nemopteridae + (Ascalaphidae + Myrmeleontidae)))) was strongly supported. In addition, E. coreanus was identified as a sister clade to the clade of (Myrmeleon formicarius + (M. immanis KJ461323 + M. immanis KM216750)).
Figure 1. Phylogenetic tree of the relationships among 21 species of Neuroptera including Euroleon coreanus. The mitochondrial genomes of 20 Neuroptera species were downloaded from GenBank and used to investigate the phylogenetic relationships, with Xanthostigma gobicola chosen as the outgroup. Numbers around the nodes show the posterior probabilities of BI (right) and the bootstrap values of ML (left). GenBank numbers of all species are shown in the figure.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. MW879174.
Additional information
Funding
References
- Aspöck U, Plant JD, Nemeschkal HL. 2001. Cladistic analysis of Neuroptera and their systematic position within Neuropterida (Insecta: Holometabola: Neuropterida: Neuroptera). Syst Entomol. 26(1):73–86.
- Beckenbach AT, Stewart JB. 2009. Insect mitochondrial genomics 3: the complete mitochondrial genome sequences of representatives from two neuropteroid orders: a dobsonfly (order Megaloptera) and a giant lacewing and an owlfly (order Neuroptera). Genome. 52(1):31–38.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Cheng CH, Gai YH, Zhang W, Shao LL, Hao JS, Yang Q. 2014. The complete mitochondrial genome of the Hybris subjacens (Neuroptera:Ascalaphidae). Mitochondrial DNA. 25(2):109–110.
- Cheng CH, Sun XY, Gai YH, Hao JS. 2015. The complete mitochondrial genome of the Epacanthaclisis banksi (Neuroptera: Myrmeleontidae). Mitochondrial DNA. 26(6):821–822.
- Gao XY, Cai YY, Yu DN, Storey KB, Zhang JY. 2018. Characteristics of the complete mitochondrial genome of Suhpalacsa longialata (Neuroptera, Ascalaphidae) and its phylogenetic implications. PeerJ. 6(1):e5914.
- Huelsenbeck JP, Ronquist F. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 17(8):754–755.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Lalitha S. 2000. Primer premier 5. Biotech Softw Internet Rep. 1(6):270–272.
- Lan XE, Chen S, Li FH, You P. 2016. The complete mitochondrial genome of Bullanga florida (Neuroptera: Myrmeleontidae). Mitochondrial DNA Part B. 1(1):632–634.
- Liu JN, Wang TH, Jia QY, Gao XH, Wan H, Sun WY, Yang XL, Bao R, Liu JZ, Yu ZJ. 2016. Characterization of the microbial communities in the ant lion Euroleon coreanus (Okamoto) (Neuroptera: Myrmeleontidae). Neotrop Entomol. 45(4):397–403.
- Machado RJP, Gillung JP, Winterton SL, Garzón-Orduña IJ, Lemmon AR, Lemmon EM, Oswald JD. 2019. Owlflies are derived antlions: anchored phylogenomics supports a new phylogeny and classification of Myrmeleontidae (Neuroptera). Syst Entomol. 44(2):418–450.
- Machado RJP, Oswald JD. 2020. Morphological phylogeny and taxonomic revision of the former antlion subtribe Periclystina (Neuroptera: Myrmeleontidae: Dendroleontinae). Zootaxa. 4796(1):1–322.
- Negrisolo E, Babbucci M, Patarnello T. 2011. The mitochondrial genome of the ascalaphid owlfly Libelloides macaronius and comparative evolutionary mitochondriomics of neuropterid insects. BMC Genom. 12:221.
- Oswald JD. 2020. Neuropterida species of the world; [accessed 2020 Nov 20]. http://lacewing.tamu.edu/
- Simon C, Buckley TR, Frati F, Stewart JB, Beckenbach AT. 2006. Incorporating molecular evolution into phylogenetic analysis, and a new compilation of conserved polymerase chain reaction primers for animal mitochondrial DNA. Annu Rev Ecol Evol Syst. 37(1):545–579.
- Song N, Li XX, Zhai Q, Bozdoğan H, Yin XM. 2019. The mitochondrial genomes of neuropteridan insects and implications for the phylogeny of Neuroptera. Genes. 10(2):108.
- Song N, Lin AL, Zhao XC. 2018. Insight into higher-level phylogeny of Neuropterida: evidence from secondary structures of mitochondrial rRNA genes and mitogenomic data. PLOS One. 13(1):e0191826.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Wang YY, Liu XY, Garzón-Orduña IJ, Winterton SL, Yan Y, Aspöck U, Aspöck H, Yang D. 2017. Mitochondrial phylogenomics illuminates the evolutionary history of Neuropterida. Cladistics. 33(6):617–636.
- Winterton SL, Hardy NB, Wiegmann BM. 2010. On wings of lace: phylogeny and Bayesian divergence time estimates of Neuropterida (Insecta) based on morphological and molecular data. Syst Entomol. 35(3):349–378.
- Winterton SL, Lemmon AR, Gillung JP, Garzon IJ, Badano D, Bakkes DK, Breitkreuz LCV, Engel MS, Lemmon EM, Liu XY, Machado RJP, et al. 2018. Evolution of lacewings and allied orders using anchored phylogenomics (Neuroptera, Megaloptera, Raphidioptera). Syst Entomol. 43(2):330–354.
- Wu JQ, Wang SZ, Yang XZ, Guo YJ, Weng XQ, Wu SQ. 2020. The complete mitochondrial genome of Myrmeleon formicarius (Neuroptera: Myrmeleontidae). Mitochondrial DNA Part B. 5(2):1798–1799.
- Yan Y, Wang YY, Liu XY, Winterton SL, Yang D. 2014. The first mitochondrial genomes of antlion (Neuroptera: Myrmeleontidae) and split-footed lacewing (Neuroptera: Nymphidae), with phylogenetic implications of Myrmeleontiformia. Int J Biol Sci. 10(8):895–908.
- Zhang J, Wang XL. 2016. The complete mitochondrial genome of Myrmeleon immanis Walker, 1853 (Neuroptera: Myrmeleontidae). Mitochondrial DNA Part A. 27(2):1439–1440.
- Zhang LL, Yang J. 2017. The mitochondrial genome of Gatzara jezoensis (Neuroptera: Myrmeleontidae) and phylogenetic analysis of Neuroptera. Biochem Syst Ecol. 71:230–235.
- Zheng Y, Liu X. 2021. The rare antlion genus Thaumatoleon Esben-Petersen, 1921 (Neuroptera: Myrmeleontidae: Nemoleontinae) newly recorded from mainland China, with first male description. Zootaxa. 4952(3):551–560.
