Abstract
Cephalopholis miniata belongs to the family Serranidae of Perciformes, which is one of the most important species in coral-reef ecosystem. In this study, its complete mitochondrial genome is presented. The assembled mitogenome is 16,585 bp and includes 13 protein-coding genes (PCGs), 22 transfer RNA genes (tRNAs), two ribosomal RNA genes (12S and 16S), and an AT-rich region (also called control region, CR). The overall base composition is 29.2% A, 26.1% T, 16.2% G, and 28.5% C, with an A + T bias of 55.3%. Except ND6 and eight tRNAs, all other mitochondrial genes are encoded on the heavy strand. Phylogenetic analysis shows that all species of the genus Cephalopholis cluster into a clade, including the newly sequenced mitogenome of C. miniata.
Groupers are bottom-associated fishes found in the tropical and subtropical waters of all oceans. The coral hind, Cephalopholis miniata (Forsskål 1775), is a kind of groupers belonging to the family Serranidae of Perciformes. It is widespread throughout the tropical waters of the Indo-West Pacific area, including Durban, South Africa, the Red Sea, and Line Islands (Rocha Citation2018). C. miniata is an important species in commercial fisheries at the local level. Meanwhile, it shows important ecological functions because it is one of the major predators feeding on a variety of fishes, crustaceans, and cephalopods in coral-reef ecosystem (Randall and Brock Citation1960; Heemstra and Randall Citation1993; Pinault et al. Citation2014). In this study, we described the complete mitochondrial genome (mitogenome) of C. miniata and explored the phylogenetic relationship within Serranidae. All the results presented in this study would provide insights into the evolution and conservation genetics of the genus Cephalopholis.
Sample of C. miniata was collected from Sansha, Hainan Province of China (112°22′40″E, 16°50′25″N) and stored in laboratory of Zhejiang Ocean University with accession number HNSS021. Total genomic DNA was extracted using a phenolch: loroform extraction protocol (Sambrook and Russell Citation2006). Based on the existing complete mitogenome of Cephalopholis urodeta (NC_030057), 12 pairs of primers were designed. The samples were amplified by PCR, and then sequenced using Sanger sequencing technology. Annotation of the complete mitogenome sequence was performed using Sequin (version 15.10, http://www.ncbi.nlm.nih.gov/Sequin/) and tRNAscan-SE 1.21 (Lowe and Chan Citation2016). The complete mitogenome sequence of C. miniata was deposited into GenBank database with the accession number MW423580.
The complete mitogenome of C. miniata was a closed-circular molecule of 16,585 bp in length. It contained 13 protein-coding genes (PCGs), two ribosomal RNA genes (12S and 16S), 22 transfer RNA genes (tRNA), and a putative control region (CR), which was the same as other Perciformes species (Ceruso et al. Citation2018; Mascolo et al. Citation2018; Ceruso et al. Citation2020). The overall base composition was A (29.2%), T (26.1%), G (16.2%), C (28.5%); respectively, with a slight AT bias (55.3%). Except one PCGs (ND6) and eight tRNAs (tRNA-Gln, Ala, Asn, Cys, Tyr, SerAGT, Glu, and Pro), which were distributed on the light (L-) strand, the rest of mitochondrial genes were distributed on the heavy (H-) strand. The total length of 13 PCGs was 11,406 bp. All of the 13 PCGs were initiated by the start codon ATN (ATA and ATG), with an exception (GTG) in COI. The majority of the 13 PCGs terminated with TAA or TAG, while three other PCGs (COII, ND4, and Cyt b) used a single T as a stop codon. The mitogenome of C. miniata contained 22 tRNA genes scattered throughout the genome. Most tRNAs displayed a typical cloverleaf secondary structure except for tRNA-Ser (GCT) that lacked the dihydrouridine (DHU) arm. The 12S and 16S rRNA genes were located between the tRNA-Phe and tRNA-Leu (TAA) genes, and were separated by the tRNA-Val gene. The CR of C. miniata was located between tRNA-Phe and tRNA-Pro, with a total length of 878 bp.
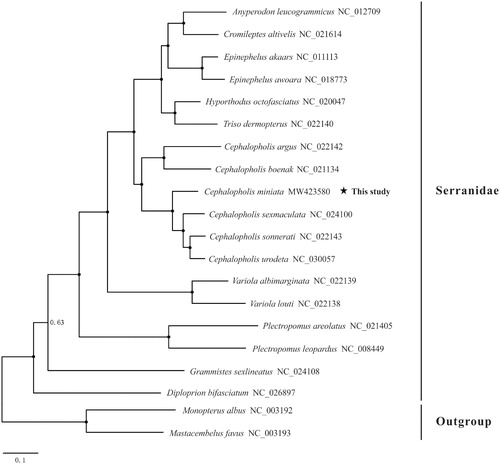
To ascertain the phylogenetic position of C. miniata, we downloaded 17 mitogenomes of Serranidae species and two outgroup species (Mastacembelus favus and Monopterus albus) from GenBank to conduct phylogenetic analysis (). The Bayesian Inference (BI) tree was constructed using the software MrBayes 3.2.6 (Ronquist et al. Citation2012) based on the nucleotide sequences of 12 PCGs (except ND6). The GTR + F + I + G4 model selected by ModelFinder (Kalyaanamoorthy et al. Citation2017) was applied in the BI analysis. The results showed that all species of the genus Cephalopholis clustered into a clade, including the newly sequenced mitogenome of C. miniata, indicating the monophyly of this genus. We expect the present results will provide an important dataset for phylogenetic and taxonomic analyses of genus Cephalopholis species.
Figure 1. Phylogenetic tree of Serranidae species inferred from the 12 PCGs based on Bayesian inference (BI) analysis. Values at the branches represent Bayesian posterior probabilities (BPP). Node marked with solid circle indicates 100% supporting value. The number after the species name is the GenBank accession number.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
Mitochondrial genome sequence can be accessed via accession number MW423580 (https://www.ncbi.nlm.nih.gov/nuccore/MW423580) in the NCBI GenBank.
Additional information
Funding
References
- Ceruso C, Venuti I, Osca D, Caputi L, Anastasio A, Crocetta F, Sordino P, Pepe T. 2020. The complete mitochondrial genome of the sharpsnout seabream Diplodus puntazzo (Perciformes: Sparidae). Mitochondrial DNA Part B. 5(3):2379–2381.
- Ceruso M, Mascolo C, Palma G, Anastasio A, Pepe T, Sordino P. 2018. The complete mitochondrial genome of the common dentex, Dentex dentex (Perciformes: Sparidae). Mitochondrial DNA Part B. 3(1):391–392.
- Heemstra PC, Randall JE. 1993. FAO Species Catalogue. Vol. 16. Groupers of the world (family Serranidae, subfamily Epinephelinae). An annotated and illustrated catalogue of the grouper, rockcod, hind, coral grouper and lyretail species known to date. FAO Fisheries Synopsis, Vol. 125. Rome: FAO; p. 49–50.
- Kalyaanamoorthy S, Minh BQ, Wong TK, von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–589.
- Lowe TM, Chan PP. 2016. tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 44(W1):W54–W57.
- Mascolo C, Ceruso M, Palma G, Anastasio A, Sordino P, Pepe T. 2018. The complete mitochondrial genome of the axillary seabream, Pagellus acarne (Perciformes: Sparidae). Mitochondrial DNA Part B. 3(1):434–435.
- Pinault M, Bissery C, Gassiole G, Magalon H, Quod J-P, Galzin R. 2014. Fish community structure in relation to environmental variation in coastal volcanic habitats. J Exp Mar Biol Ecol. 460:62–71.
- Randall JE, Brock VE. 1960. Observations on the ecology of epinepheline and lutjanid fishes of the Society Islands, with emphasis on food habits. Trans Am Fish Soc. 89(1):9–16.
- Rocha LA. 2018. Cephalopholis miniata. IUCN Red List of Threatened Species. e.T132732A100455926.
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Hohna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542.
- Sambrook J, Russell DW. 2006. Purification of nucleic acids by extraction with phenol: chloroform. Cold Spring Harb Protoc. 2006(1):pdb.prot4455.
