Abstract
Verbena officinalis is one kind of traditional medical herb which has potential for multiple diseases’ treatment. In this study, the complete chloroplast genome sequence of V. officinalis was assembled. Its complete circular chloroplast DNA length was 153,491 bp. The genome was made up of a large single-copy region of 84,518 bp, a small single-copy region of 17,357 bp, and a pair of inverted repeat regions of 25,808 bp. The genome totally encoded 128 genes, containing 83 protein-coding genes, 37 tRNA genes, and eight rRNA genes. Phylogenetic analysis indicates that V. officinalis belongs to the verbenaceae family.
Verbena officinalis is a perennial herb native to Europe, and now has been widely cultivated in other areas of the world. It has attracted much attention because of its good medicinal value. The extract of V. officinalis showed the anti-tumor effect for H22 hepatoma ascites mice (Kou et al. Citation2013). It also had antioxidant and antifungal activities (Casanova et al. Citation2008). Moreover, it was beneficial in the aspects of neuroprotection (Lai et al. Citation2006) and anti-inflammatory activity (Calvo Citation2006). The complete chloroplast genome of V. officinalis was reported herein, which can provide a deeper understanding for its phylogeny and taxonomy.
The plant sample of V. officinalis was collected from the herb nursery of Xianyang (108.69E, 34.35 N), Shaanxi Province, China. The voucher specimen was deposited in Herbarium of the Microbiology Institute of Shaanxi, Microbiology Institute of Shaanxi, China (http://sxim.xab.cas.cn/, Yan Wang, [email protected]) under the voucher number zw2020006. The DNA from fresh leaves was extracted by CTAB method (Porebski et al. Citation1997). The DNA insert fragments about 400 bp in length were used for library construction. Based on the Illumina Novaseq Platform at Personal Biotechnology Co. Ltd (Shanghai, China) and 2 × 250 bp pair-end sequencing mode, total 5,778,366 reads of 1.45 Gbp sequencing data was generated. The cleaned reads were assembled to form contig via NOVOPlasty (v 4.2, Dierckxsens et al. Citation2017) by Aloysia citrodora as the reference chloroplast genome (NCBI accession number: NC_034695.1). The final chloroplast genome was annotated by CPGAVAS2 (Shi et al. Citation2019), and all introns/exons of genes were checked artificially.
Using the next generation sequencing technology, we assembled a circular complete chloroplast genome of V. officinalis. The genome sequence and all gene annotations were submitted to the NCBI database with accession number of MW328640. The complete chloroplast genome sequence was 153,491 bp in length. The genome was consisted of a large single-copy region (LSC, 84,518 bp), a small single-copy region (SSC, 17,357 bp) and two inverted repeat regions (IR, 25,808 bp). The whole genome encoded 128 genes, including 83 protein-coding genes, 37 tRNA genes and 8 rRNA genes.
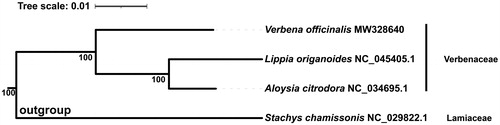
To determine the evolutionary relationship of V. officinalis, a maximum-likelihood phylogenetic tree was constructed based on 4 complete chloroplast genome sequences (). The species of Stachys chamissonis which belongs to Lamiaceae family was as outgroup. All genome sequences were retrieved from the GenBank database. These sequences were aligned using MAFFT (v 7.407, Katoh and Standley Citation2013). Then, trimAl (v 1.4.1, Capella-Gutiérrez et al. Citation2009) was applied to remove poorly-aligned and divergent regions with algorithm automated1. The remaining sequences were used to infer the phylogenetic position using IQ-TREE (v 1.6.12, Nguyen et al. Citation2015) under parameters “-nt AUTO -m MFP -bb 1000 -bnni”. The TVM + F+R3 model was chosen according to BIC and 1000 bootstrap replicates were used. The result showed that V. officinalis belong to the Verbenaceae family.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under the accession no. MW328640. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA682879, SRR13205890, and SAMN17013277 respectively.
Additional information
Funding
References
- Calvo MI. 2006. Anti-inflammatory and analgesic activity of the topical preparation of Verbena officinalis L. J Ethnopharmacol. 107(3):380–382.
- Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 25(15):1972–1973.
- Casanova E, García-Mina JM, Calvo MI. 2008. Antioxidant and antifungal activity of Verbena officinalis L. leaves. Plant Foods Hum Nutr. 63(3):93–97.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kou WZ, Yang J, Yang QH, et al. 2013. Study on in-vivo anti-tumor activity of Verbena officinalis extract. Afr J Tradit Complement Altern Med. 10(3):512–517.
- Lai SW, Yu MS, Yuen WH, et al. 2006. Novel neuroprotective effects of the aqueous extracts from Verbena officinalis Linn. Neuropharmacology. 50(6):641–650.
- Nguyen LT, Schmidt HA, von Haeseler A, et al. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Porebski S, Bailey LG, Baum BR. 1997. Modification of a CTAB DNA extraction protocol for plants containing high polysaccharide and polyphenol components. Plant Mol Biol Rep. 15(1):8–15.
- Shi L, Chen H, Jiang M, et al. 2019. CPGAVAS2, an integrated plastome sequence annotator and analyzer. Nucleic Acids Res. 47(W1):W65–W73.