Abstract
With about 153 species, the genus Androsace (Primulaceae) is known for its horticultural and economic importance. In this study, we report the complete chloroplast genome of Androsace erecta Maximowicz, a morphologically distinct species of Sect. Orthocaulon native to the Western China. The plastome of A. erecta is highly conserved in genome size, structure, and content when compared to all previously published plastomes of the genus. The phylogenomic analysis strongly supported A. erecta as sister to a clade comprising species of Sections Aizoideia and Chamaejasme. Lastly, we selected the four most variable regions across the Androsace species plastomes (trnKUUU-rps16, trnSGCU-trnGUCC, psbE-petL, and infA-rps8), which were considered to be suitable candidate DNA barcodes for Androsace.
Plants of the genus Androsace are highly prized for their ornamental and pharmaceutical values (Smith et al. Citation1997). The genus is commonly found in northern temperate regions, yet occupying a wide range of elevational gradients, from 800 to 6350 m a.s.l. (Hu and Kelso Citation1996; Dentant Citation2018). The genus is known for its morphological diversity, comprising seven highly variable sections (Hu Citation1994; Hu and Kelso Citation1996). In particular, the monotypic Section Orthocaulon contains only one species, Androsace erecta Maximowicz, whose peculiar morphology (a densely leafy stem and lack of a basal rosette) has attracted significant attention for over a century (Knuth and Pax Citation1905; Hu and Kelso Citation1996; Schneeweiss et al. Citation2004). In this study, we sequenced the complete chloroplast genome of A. erecta for the first time. To investigate its phylogenetic position, we selected another five species (see ), which altogether represent the four major sections of Androsace, including Sect. Chaemajasme, the sister clade of Sect. Orthocaulon. Subsequent phylogenomic analyses were conducted based on plastome data available in GenBank and the newly generated plastome built by us. The sample of A. erecta was collected from a wild population found in Deqin County, near the National Highway G214 (28.451833°N, 98.858769°E). The voucher was deposited at the Herbarium of Xishuangbanna Tropical Botanical Garden, Chinese Academy of Sciences (V000018, HITBC, http://hitbc.xtbg.ac.cn/, [email protected]).
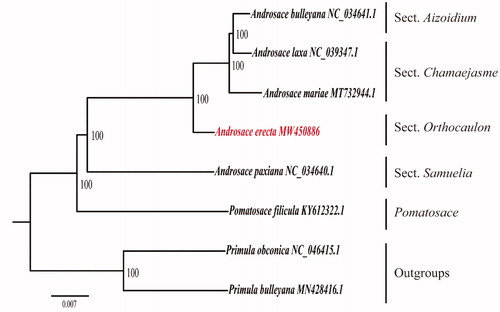
Figure 1. Maximum likelihood tree of 8 species based on the complete chloroplast genome data. The branch statistics are the bootstrap values.
Total genomic DNA was extracted from leaves dried with silica gel, using a modified CTAB procedure (Doyle and Doyle Citation1987). About 8 Gb of genomic data were generated by an Illumina NovaSeq 6000 platform with a reading length of 150 bp. More than 28 million clean reads were de novo assembled and annotated by the GetOrganelle toolkit (Jin et al. Citation2020) and CPGAVAS2 (Shi et al. Citation2019), and then manually adjusted in Geneious (Kearse et al. Citation2012). The complete chloroplast genome (over 133× coverage) was aligned by MAFFT (Katoh and Standley Citation2013). The resulting genome matrix was subsequently used to reconstruct the maximum likelihood tree through RAxML under a GTRGAMMA + I model with 1000 bootstraps (Stamatakis Citation2014) on the CIPRES online portal (https://www.phylo.org/). The tree was visualized with FigTree version 1.4.4 (Rambaut Citation2010). The nucleotide diversity (π) of the five Androsace plastomes were calculated by DnaSP with 500 bp window size and 200 bp step size respectively (Rozas et al. Citation2017).
With a GC content of 37.2%, the plastome of A. erecta was a circular DNA molecule of 153,547 bp (MW450886), with a typical quadripartite structure including two IRs of 26,008 bp separating the LSC of 83,745 bp and the SSC of 17,786 bp. We identified 111 unique genes, 80 CDS (coding sequences), 34 transfer RNAs (tRNAs), and four ribosomal RNAs (rRNAs), which are all consistent with previously published Androsace plastomes. Unlike its unique morphology, the plastome of A. erecta is highly conserved in genome size, structure, and gene content, similar to most Primulaceae taxa.
As rooted by two Primula species (), our phylogenetic analysis confirmed the previous infrageneric scheme of Androsace at the sectional level (Schneeweiss et al. Citation2004; Wang et al. Citation2004), and strongly supported that Sect. Orthocaulon Hand. - Mazz. (A. erecta) is sister to the clade comprising Sect. Chamaejasme Koch. (A. laxa, A. mariae) and Sect. Aizoidium Hand.- Mazz. (A. bulleyana). However, the genus Pomatoace, which is nested in Androsace in previous studies (Boucher et al. Citation2012; Roquet et al. Citation2013), was found to be the sister clade of the Androsace with a high support value (BS: 100). Considering our limited taxon sampling, the result still needs further investigation.
As a popular garden plant (Smith et al. Citation1997), correct species identification of Androsace is important for trading the species (Dixon et al. Citation2016; Dentant Citation2018). Therefore, we have evaluated and recognized four highly variable regions (trnKUUU-rps16, trnSGCU-trnGUCC, psbE-petL, infA-rps8; pi > .08) by comparing the nucleotide diversity of the plastomes of the five species considered here. They are considered to have a high potential for developing DNA barcodes and as genetic markers for ecological and evolutionary studies of Androsace (Shaw et al. Citation2007; Dong et al. Citation2012).
Acknowledgments
We are grateful for the support from the HPC Platform of the Public Technology Service Center, Xishuangbanna Tropical Botanical Garden, Chinese Academy of Sciences.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the finding of this study are openly available in GenBank at https://www.ncbi.nlm.nih.gov/nuccore/MW450886 under the accession number MW450886. The associated BioProject, BioSample, and SRA numbers are PRJNA706062, SAMN18115972, and SUB9180715, respectively.
Additional information
Funding
References
- Boucher FC, Thuiller W, Roquet C, Douzet R, Aubert S, Alvarez N, Lavergne S. 2012. Reconstructing the origins of high-alpine niches and cushion life form in the genus Androsace s.l. (Primulaceae). Evolution. 66(4):1255–1268.
- Dentant C. 2018. The highest vascular plants on Earth. Alp Bot. 128(2):97–106.
- Dentant C, Lavergne S, Malecot V. 2018. Taxonomic revision of West-Alpine cushion plant species belonging to Androsace subsect Aretia. Bot Lett. 165(3–4):337–351.
- Dixon CJ, Gutermann W, Schönswetter P, Schneeweiss GM. 2016. Taxonomy and nomenclature of the polymorphic European high mountain species Androsace vitaliana (L.) Lapeyr. (Primulaceae). PhytoKeys. 75:93–106.
- Dong W, Liu J, Yu J, Wang L, Zhou S. 2012. Highly variable chloroplast markers for evaluating plant phylogeny at low taxonomic levels and for DNA barcoding. PLoS One. 7(4):e35071.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19(1):11–15.
- Hu Q. 1994. On the geographical distribution of the Primulaceae. J Trop Subtrop Bot. 2(4):1–14.
- Hu CM, Kelso S. 1996. Flora of China. 15. Cambridge: Science Press, Beijing & Cambridge University Press; p. 295.
- Jin JJ, Yu WB, Yang JB, Song Y, dePamphilis CW, Yi TS, Li DZ. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):241.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Knuth R, Pax F. 1905. Das Pflanzenreich. 4. Leipzig, Germany: Wilhelm Engelmann; p. 386.
- Rambaut A. 2010. FigTree v1.3.1. Edinburgh, Scotland: Institute of Evolutionary Biology, University of Edinburgh. http://tree.bio.ed.ac.uk/software/figtree/.
- Roquet C, Boucher FC, Thuiller W, Lavergne S. 2013. Replicated radiations of the alpine genus Androsace (Primulaceae) driven by range expansion and convergent key innovations. J Biogeogr. 40(10):1874–1886.
- Rozas J, Ferrer-Mata A, Sánchez-DelBarrio JC, Guirao-Rico S, Librado P, Ramos-Onsins SE, Sánchez-Gracia A. 2017. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol Biol Evol. 34(12):3299–3302.
- Schneeweiss GM, Schonswetter P, Kelso S, Niklfeld H. 2004. Complex biogeographic patterns in Androsace (Primulaceae) and related genera: evidence from phylogenetic analyses of nuclear internal transcribed spacer and plastid trnL-F sequences. Syst Biol. 53(6):856–876.
- Shaw J, Lickey EB, Schilling EE, Small RL. 2007. Comparison of whole chloroplast genome sequences to choose noncoding regions for phylogenetic studies in angiosperms: the tortoise and the hare III. Am J Bot. 94(3):275–288.
- Shi L, Chen H, Jiang M, Wang L, Wu X, Huang L, Liu C. 2019. CPGAVAS2, an integrated plastome sequence annotator and analyzer. Nucleic Acids Res. 47(W1):W65–W73.
- Smith G, Lowe D, Christopher GW. 1997. The genus ‘Androsace’: a monograph for gardeners and botanists. Dorset: Alpine Garden Society Publications Limited,
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Wang YJ, Li XJ, Liu JQ. 2004. Molecular phylogeny and biogeography of Androsace (Primulaceae) and the convergent evolution of cushion morphology. J Syst Evol. 42(6):481–499.
