Abstract
The complete mitochondrial genome of Aleochara postica Walker, 1858 was determined in this study. It is 15,473 bps in length, containing 13 protein-coding genes, 22 tRNA genes, 2 rRNA genes, and a 778 bp A + T-rich control region. Most PCGs use the conventional ATN start codon, except for cox1 and nad1. Two genes (cox1 and cox3) use single T residue as stop codon rather than the routinely used TAA or TAG. All tRNAs, except for TrnS1, exhibit the cloverleaf secondary structure. ML phylogenetic analysis using 13 PCGs of 52 beetle species indicated that A. postica was clustered with other members of the subfamily Aleocharinae as conventional taxonomy predicted.
Aleochara postica Walker, Citation1858, a rove beetle species, belongs to the genus Aleochara Gravenhorst (Staphylinidae: Aleocharinae: Aleocharini). Aleochara with more than 500 species distributed worldwide is not only one of the most speciose genera in Aleocharinae, but also a distinctive group in lifestyle (Klimaszewski Citation1984; Caron et al. Citation2019). Most of its species have their larvae specifically feeding on pupae of cyclorrhaphous Diptera as ectoparasitoids, and adult beetles preying on dipteran eggs and larvae (Luo and Zhou Citation2012; Yamamoto and Maruyama Citation2016; Caron et al. Citation2019). Thus, they could be utilized for biological control against noxious flies (Yamamoto and Maruyama Citation2016; Caron et al. Citation2019). Nevertheless, no complete mitogenome of Aleochara was available so for. In compensation, we present herein the complete mitogenome of A. postica, which is widely distributed in China, Japan, Korea, Sri Lanka, and much of Oriental region (Yamamoto and Maruyama Citation2016). In our study, the adults were collected in 2020, from Guiyang Huaxi District (26°23'06″N, 106°36'56″E, 1163 m), Guizhou, China, using maggoty dead fish bait.
The high-throughput sequencing was performed at Sangon Biotech (Shanghai) Co., Ltd., China, using Illumina HiSeq2500 platform (Illumina, San Diego, CA). The de novo assembly of mitogenome and correctness check were carried out with the software combination of SPAdes V.3.14.1 (Bankevich et al. Citation2012), MitoZ V.2.3 (Meng et al. Citation2019), and Pilon V.1.23 (Walker et al. Citation2014). MITOS Web Server (http://mitos2.bioinf.uni-leipzig.de/index.py) was utilized for annotation. The remaining alcohol-preserved specimen tissue and the total DNA after sequencing were deposited under −20 °C in the Insect Collection of Guizhou University of Traditional Chinese Medicine, Guiyang, China (Yanpeng Cai, [email protected], Voucher specimens: GZUTCM:002).
The complete circular mitogenome of A. postica (GenBank: MW284907) is 15,473 bps in length, containing the typical metazoan mitochondrial genes (13 protein-coding genes, 22 tRNA genes, 2 rRNA genes) and a 778 bp long A + T-rich control region. Most PCGs of A. postica use conventional start codons (ATN) and stop codons (TAA or TAG). Whereas, cox1 and nad1 genes initiate with putative start codons GAT and TTG respectively, cox1 and cox3 genes use single T as incomplete stop codon. Twenty-one out of 22 tRNAs exhibit the typical clover-leaf structure. TrnS1 as the only exception lacks the DHU arm, and that the anticodon of trnS1 is UCU instead of routinely used GCU.
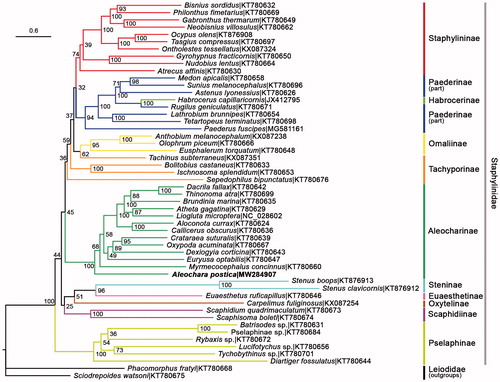
The ML phylogenetic tree was reconstructed using IQTREE V.2.07 (Nguyen et al. Citation2015) for family Staphylinidae, based on 13 PCGs of A. postica plus 51 species obtained from GenBank, among which two representatives of Leiodidae were selected as outgroups (Lin et al. Citation2018). The partitioning scheme for the three codon positions of the 13 genes was determined by the TESTMERGE option in IQTREE. Ten partitions were finally divided and allocated with their own best fit substitution model and parameters (GTR + F + I + G4, GTR + F + I + G4, GTR + F + I + G4, TN + F+G4, GTR + F + I + G4, TVM + F + I + G4, TN + F+G4, GTR + F + I + G4, K3Pu + F + I + G4, TN + F + I + G4). 1000 replicates of standard bootstrap analysis were executed to produce the bootstrap support values. As a result, seven subfamilies with multiple available representatives (Aleocharinae, Omaliinae, Paederinae, Pselaphinae, Scaphidiinae, Staphylininae, Steninae) were recovered as monophyla. Paederinae was a sibling group to Staphylininae which was supported by multiple previous studies (e.g. Mckenna et al. Citation2015; Tihelka et al. Citation2020). Tachyporinae was polyphyletic, which was proved in Yamamoto (Citation2021) as well. The only representative of Habrocerinae was oddly nested in Peaderinae. Aleochara postica was clustered in Aleocharinae as morphological taxonomy predicted ().
Disclosure statement
No potential conflict of interest was reported by the author.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/nuccore/MW284907 under the Accession no. MW284907. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA728529, SRR14508658, and SAMN19091164, respectively.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Caron E, Moussallem M, Bortoluzzi S. 2019. Revision of Brazilian species of Aleochara Gravenhorst of the subgenus Aleochara (Coleoptera: Staphylinidae: Aleocharinae). Zootaxa. 4712(1):1–033.
- Klimaszewski J. 1984. A revision of the genus Aleochara Gravenhorst of America north of Mexico. Mem Entomol Soc Can. 116(S129):3–129.
- Lin AL, Song N, Zhao XC, Zhang FM. 2018. Analysis of the nearly complete mitochondrial genome of Paederus fuscipes (Coleoptera: Staphylinidae). Mitochondrial DNA Part B. 3(1):85–87.
- Luo TH, Zhou HZ. 2012. Taxonomic study of the subgenus Aleochara (s. str.) Gravenhorst (Coleoptera: Staphylinidae: Aleocharinae) in China, with descriptions of four new species. *Ann Entom Soc Am. 105(2):179–200.
- McKenna DD, Farrell BD, Caterino MS, Farnum CW, Hawks DC, Maddison DR, Seago AE, Short AEZ, Newton AF, Thayer MK. 2015. Phylogeny and evolution of Staphyliniformia and Scarabaeiformia: forest litter as a stepping stone for diversification of nonphytophagous beetles. Syst Entomol. 40(1):35–60.
- Meng GL, Li YY, Yang CT, Liu SL. 2019. MitoZ: a toolkit for animal mitochondrial genome assembly, annotation and visualization. Nucleic Acids Res. 47(11):e63.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Tihelka E, Thayer MK, Newton AF, Cai CY. 2020. New data, old story: molecular data illuminate the tribal relationships among rove beetles of the subfamily Staphylininae (Coleoptera: Staphylinidae). Insects. 11(3):164.
- Walker F. 1858. Characters of some apparently undescribed Ceylon Insects. Ann Mag Nat Hist. 2(9):202–209.
- Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, Sakthikumar S, Cuomo CA, Zeng Q, Wortman J, Young SK, et al. 2014. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One. 9(11):e112963.
- Yamamoto SH. 2021. Tachyporinae revisited: phylogeny, evolution, and higher classification based on morphology, with recognition of a new rove beetle subfamily (Coleoptera: Staphylinidae). Biology. 10(4):323.
- Yamamoto SH, Maruyama MT. 2016. Revision of the subgenus Aleochara Gravenhorst of the parasitoid rove beetle genus Aleochara Gravenhorst of Japan (Coleoptera: Staphylinidae: Aleocharinae). Zootaxa. 4101(1):1–068.