Abstract
Cornus alba has been used as antiphlogistic, hemostatic and diuretic treatments in Korea, and which is one of the most important ornamental shrubs in China. Here, we reported the complete chloroplast genome sequence of C. alba. The size of the chloroplast genome is 158,451 bp in length, including a large single copy region (LSC) of 87,778 bp, a small single copy region (SSC) of 18,927 bp, and a pair of inverted repeated regions of 25,873 bp. The C. alba chloroplast genome encodes 132 genes, including 85 protein coding genes, 38 tRNA genes, and 8 rRNA genes. Phylogenetic tree showed that C. alba with the species of C. macrophylla and C. sanguinea formed a strongly supported clade.
The genus Cornus Linnaeus (Cornaceae) consists of more than 55 species, mostly shrubs and small trees (Bjorøy et al. Citation2007). Cornus alba Linn., also known as red-barked or Siberian dogwood, is native to Siberia, northern China and Korea (Park et al. Citation2016). The stems and leaves of C. alba have been used as antiphlogistic, hemostatic and diuretic treatments in Korea. This plant is one of the most important ornamental shrub in China due to its colorful autumn leaves, its crimson bark in the winter season, white flowers and its berries with a bluish white metallic shine (Luo Citation2020). The advent of high-throughput sequencing technology made it possible to obtain large amounts of genomic data rapidly (He, Yuan, et al. Citation2017). In this study, we reported the complete chloroplast (cp) genome of C. alba (Genbank accession no. MW368898) to provide genomic and genetic information for further research.
The fresh leaves of C. alba were collected from the campus of Henan University, China (34°49'19.68"N, 114°18'51.32"E), and a voucher specimen was kept at the Herbarium of Henan University (Voucher number: Wangjun Yuan 2020080601, Kaifeng, Henan Province, China, Wangjun Yuan and [email protected]). Total genomic DNA was extracted with the modified SDS method. Sequencing libraries were generated using NEBNext® Ultra™ DNA Library Prep Kit for Illumina (NEB, Bloomington, IN) following manufacturer’s recommendations, which were sequenced using Illumina NovaSeq PE150 at the Beijing Novogene Bioinformatics Technology Co., Ltd. (Beijing, China). Approximately 5 Gb raw data were generated, and these were trimmed and assembled into contigs using a CLC Genomics Workbench 9.5.2 (CLC Inc., Aarhus, Denmark) with the chloroplast genome sequence of C. macrophylla (GenBank accession no. NC044810) as a reference. The complete cp genome was assembled and annotated using Geneious R11 (Biomatters, Auckland, New Zealand) following description (He, Liu, et al. Citation2017; Liu et al. Citation2018).
The chloroplast genome of C. alba was 158,451 bp in length, containing a large single copy region (LSC) of 87,778 bp, a small single copy region (SSC) of 18,927 bp, and a pair of inverted repeat (IR) regions of 25,873 bp. Genome annotation predicted 132 genes, including 85 protein coding genes, 38 tRNA genes, and 8 rRNA genes. The overall GC-content of the chloroplast genome was 37.9%, and the corresponding values in the LSC, SSC, and IR regions were 36.0%, 32.2%, and 43.1%, respectively.
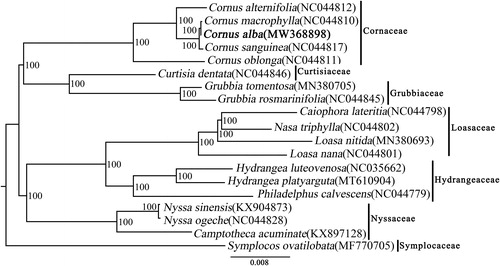
The phylogenetic tree including C. alba and other 18 species was constructed by whole cp genome sequences using the maximum-likelihood(ML) method implemented in RAxML-HPC v8.1.11 on the CIPRES cluster , which showed that C. alba with the species of C. macrophylla and C. sanguinea formed a clade with a 100% bootstrap value (). The topology of Cornales is identical to that of Xiang et al. (Citation2011).
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under the accession no. MW368898. The associated ﹡BioProject﹡, ﹡SRA﹡, and﹡Bio-Sample﹡ numbers of the raw sequence data and the genome are PRJNA720529, SRR14181230, and SAMN18653152, respectively.
Additional information
Funding
References
- Bjorøy Ø, Fossen T, Andersen ØM. 2007. Anthocyanin 3-galactosides from Cornus alba 'Sibirica’ with glucosidation of the B-ring. Phytochemistry. 68(5):640–645.
- He Y, Yuan W, Dong M, Han Y, Shang F. 2017. The first genetic map in sweet osmanthus (Osmanthus fragrans lour.) using specific locus amplified fragment sequencing. Front Plant Sci. 8:1621.
- He YX, Liu LX, Yang SH, Dong MF, Yuan WJ, Shang FD. 2017. Characterization of the complete chloroplast genome of Chinese fringetree (Chionanthus retusus). Conserv Genet Resour. 9(3):1–4.
- Liu LX, Li P, Zhang HW, Worth JRP. 2018. Whole chloroplast genome sequences of the Japanese hemlocks, Tsuga diversifolia and T. sieboldii, and development of chloroplast microsatellite markers applicable to East Asian Tsuga. J Forest Res. 23(5):318–323.
- Luo QQ. 2020. Molecular identification of collect otrichum species causing anthracnose disease of Cornus alba “Bud′s Yellow. Plant Protect Chin. 46(5):102–109.
- Park KH, Yin J, Yoon KH, Hwang YJ, Min WL. 2016. Antiproliferative effects of new dimeric ellagitannin from Cornus alba in prostate cancer cells including apoptosis-related S-phase arrest. Molecules. 21(2):137.
- Xiang QYJ, Thomas DT, Xiang QP. 2011. Resolving and dating the phylogeny of Cornales – effects of taxon sampling, data partitions, and fossil calibrations. Mol Phylogenet Evol. 59(1):123–138.