Abstract
Land leeches of genus Haemadipsa (Family Haemadipsidae) are widely distributed in South East Asia. Haemadipsa crenata Ngamprasertwong is a blood-feeding species firstly reported from Thailand. A complete mitochondrial genome of H. crenata was characterized in this study for further genetic exploration on land leech. The reads were assembled into a circular mitogenome of 14,725 bp in length. The AT content of H. crenata mitogenome is 76.79%. The annotated mitogenome contains 22 tRNAs, 2 rRNAs, and 13 protein-coding genes (PCGs), and the structure of PCG open reading frames was confirmed. Finally, the phylogenetic relationship of H. crenata and other leech species were reconstructed using mitogenomes.
Leeches (Annelida: Hirudinea) are a type of hermaphroditic and carnivorous worms with a constant number of segments and two distinctive locomotive suckers. Land leeches of family Haemadipsidae are distributed in some tropical areas of Asia and Australia (Borda et al. Citation2008). According to previous records, genus Haemadipsa (Haemadipsidae) is prevalent in Southeast Asia (Ngamprasertwong and Panha Citation2007). Haemadipsa crenata was first identified and described as a novel species in Thailand in 2007 (Ngamprasertwong and Panha Citation2007). The previously published studies of phylogenetic relationship between land leech species, including Haemadipsa crenata, were based on short barcode sequences like 18S, 28S or COI (Siddall and Burreson Citation1998; Borda and Siddall Citation2010; Schnell et al. Citation2018). Therefore, the phylogenetic relationship of land leech species remains worth of further exploration. Here, a complete mitogenome of the land leech H. crenata was presented. This mitogenome should facilitate further study on this issue.
The living H. crenata samples were captured from Mengsong Village, Menglong Town, Jinghong City, Yunnan Province, China (N21°53′, E100°63′). The leeches were deposited into pure ethyl alcohol and brought back to the lab. The jaw of leech was used for raw DNA materials extraction using CTAB method following a previous study (Skevington and Yeates Citation2000). The library construction and sequencing procedure followed the standard protocol of Illumina (Illumina Inc., San Diego, CA, USA). Then, the qualified DNA library (PE150) was sequenced on Illumina NovaSeq platform.
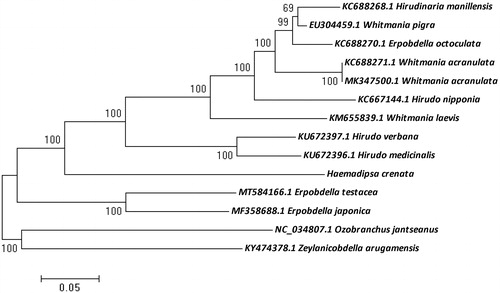
In total, 4.5 Gb data was generated and deposited into the NCBI database (BioProject: PRJNA695059, GenBank: MW711186). Voucher specimen was labeled with a unique serial number (CSU-KMU-MG20201101-1), then deposited into herbarium of department of College of Agriculture and Life Sciences, Kunming University. The cleaned reads were assembled into a 14,725-bp complete mitochondrial genome using SPAdes (Bankevich et al. Citation2012). In total, 13 protein coding-genes were identified from the mitogenome annotated with Mitochondrial Genome annotation (MITOS2) (Bernt et al. Citation2013) and open reading frames of each gene were confirmed by ORFfinder (https://www.ncbi.nlm.nih.gov/orffinder/). The tRNA secondary structures were compared with other leech tRNA sequences. 2 rRNA genes were also annotated in leech genome. Before the phylogenetic analysis, the mito-genome sequence of H. crenata was aligned with mtDNA sequences of 13 other leech species from the GenBank using the ClusterW (gap opening penalty of 30) embedded in the MEGA 7 software. Phylogenetic relationship was analyzed based on 13 mitogenomic protein coding sequences of H. crenata and other leeches using maximum likelihood (ML) method in MEGA 7 software with bootstrap of 1000 bootstrap replicates (Kumar et al. Citation2016). Ozobranchus jantseanus and Zeylanicobdella arugamensis were rooted as outgroups (). The phylogenetic tree indicated that H. crenata represents a basal branch separated from the other Hirudinea species, and the relationship between those Hirudinea leeches was similar to previous researches (Nikitina et al. Citation2016; Liu et al. Citation2017).
Disclosure statement
All authors announce no conflicts of interest in present paper.
Data availability statement
The data that support the findings of this study are now available. (https://www.ncbi.nlm.nih.gov/sra/PRJNA695059)
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Borda E, Oceguera-Figueroa A, Siddall ME. 2008. On the classification, evolution and biogeography of terrestrial haemadipsoid leeches (Hirudinida: Arhynchobdellida: Hirudiniformes). Mol Phylogenet Evol. 46(1):142–154.
- Borda E, Siddall ME. 2010. SME. Insights into the evolutionary history of Indo-Pacific bloodfeeding terrestrial leeches (Hirudinida:Arhynchobdellida:Haemadipisdae). Invert Syst. 24(5):456–472.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for Bigger Datasets. Mol Biol Evol. 33(7):1870–1874.
- Liu X, Luo D, Zhao Y, Zhang Q, Zhang J. 2017. Complete mitochondrial genome of Ozobranchus jantseanus (Hirudinida: Arhychobdellida: Ozobranchidae). Mitochondrial DNA Part B. 2(1):232–233.
- Ngamprasertwong TT, Panha S. 2007. Two new species of land leeches from Thailand (Hirudiniformes: Haemadipsidae). Nat Hist J Chulalongkorn Univ. 7:155–159.
- Nikitina A, Babenko V, Akopian T, Shirokov D, Manuvera V, Kurdyumov A, Kostryukova E, Lazarev V. 2016. Draft mitochondrial genomes of Hirudo medicinalis and Hirudo verbana (Annelida, Hirudinea). Mitochondrial DNA Part B. 1(1):254–256.
- Schnell IB, Bohmann K, Schultze SE, Richter SR, Murray DC, Sinding M-HS, Bass D, Cadle JE, Campbell MJ, Dolch R, et al. 2018. Debugging diversity: a pan-continental exploration of the potential of terrestrial blood-feeding leeches as a vertebrate monitoring tool. Mol Ecol Resour. 18(6):1282–1298.
- Siddall ME, Burreson EM. 1998. Phylogeny of leeches (Hirudinea) based on mitochondrial cytochrome c oxidase subunit I. Mol Phylogenet Evol. 9(1):156–162.
- Skevington JH, Yeates DK. 2000. Phylogeny of the syrphoidea (Diptera) inferred from mtDNA sequences and morphology with particular reference to classification of the pipunculidae (Diptera). Mol Phylogenet Evol. 16(2):212–224.