Abstract
Veratrum oxysepalum Turcz. is a medicinal plant belonging to Melanthiaceae occurring in Northeast China. However, there are still limited genomic resources available for genus Veratrum. The complete chloroplast (cp) genome of V. oxysepalum was determined and analyzed in this study. The complete cp genome was 153,705 bp. That contains a large single copy (LSC) region of 83,384 bp, a small single copy (SSC) region of 17,607 bp, which were separated by a pair of 26,358 bp inverted repeat regions (IRs). A total of 135 genes were annotated, including 83 protein-coding genes, 38 tRNAs, and eight rRNAs. Phylogenetic analysis using total chloroplast genome sequence of 21 species revealed that V. oxysepalum was closely relates to V. patulum of Veratrum with 100% bootstrap value.
There are approximately 17–45 species of perennial herbaceous plants of the genus Veratrum L. in the world. (Zomlefer et al. Citation2003). Veratrum oxysepalum Turcz., a perennial herb belonging to the genus Veratrum. It has widely distributed in hillside forest and wet meadow in Liaoning, Jilin and Heilongjiang provinces of China (Editorial Committee of Chinese Academy of Sciences Citation1980). V. oxysepalum was usually treated as traditional Chinese herbal medicines, its roots and rhizomes used to treat aphasia symptoms arising from apoplexy, wind type dysentery, jaundice, head-ache, scabies, chronic malaria and other disorders (Cong et al. Citation2020). In addition, genus Veratrum contains toxic compounds such as ester-alkaloids, there are numerous cases of mistaken identity that have resulted in Veratrum poisoning (Christopher and McDougal Citation2014). For the safety of medication, it is necessary to accurately identify the species Veratrum. Here, we reported the complete chloroplast genome of V. oxysepalum to provide genomic resource for further conservation genetics and phylogenetic analysis.
The plant sample were collected from Jilin (43°24′N, 126°38′E), Jilin Province of China, and voucher specimens (220007009) were deposited in Herbarium of Yunnan University of Chinese Medicine. Firstly, total genomic DNA was isolated from liquid nitrogen frozen and ground leaf material using the plant DNA extraction kit (Bioteke Corporation, China) and sequencing was performed on the Illumina HiSeq 2500 platform (Illumina, San Diego, CA). Secondly, we assembled the clean data of 3 Gb based on NOVOPlasty (Dierckxsens et al. Citation2017). The assembled complete chloroplast genome was annotated with the online annotation tool DOGMA (Wyman et al., Citation2004), and corrected manually with the Geneious R11 11.1.5 (Biomatters Ltd., Auckland, New Zealand).
The circular chloroplast genome of V. oxysepalum was 153,705 bp (GenBank accession number: MW147219) in length, contained a large single-copy (LSC) region of 83,384 bp, a small single-copy (SSC) region of 17,607 bp, separated by a pair of inverted repeat (IR) regions of 26,358 bp each. A total of 135 genes were annotated, including 83 protein-coding genes, 38 tRNAs, and eight rRNAs. The overall GC content of the cp genome, LSC, SSC and IR regions are 37.7%, 35.7%, 31.4%, 42.9%, respectively.
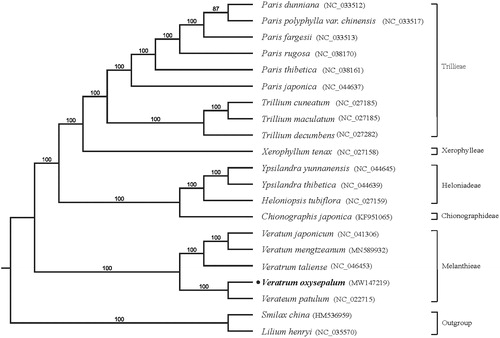
To further investigate its phylogenetic position of V. oxysepalum, plastome of 21 representative species were downloaded from NCBI GenBank database, Smilax china and Lilium henryi were selected as the outgroup to construct the plastome phylogeny. All of genomes were fully aligned using MAFFT (Katoh and Standley Citation2013). The phylogeny was using with RAxML (Stamatakis Citation2014), bootstrap probability values were calculated from 1000 replicates for supporting the branches evaluated under the GTR model. The phylogenetic tree shows that V. oxysepalum and V. patulum from Melanthiaceae formed a monophyletic clade with 100% bootstrap value (). In conclusion, this study identified unique characteristics of the V. oxysepalum cp genome providing valuable information for further investigations on species identification and the phylogenetic evolution between V. oxysepalum and related species.
Disclosure statement
The authors declare that there is no conflict of interest regarding the publication of this article. The authors alone are responsible for the content and writing of the paper.
Data availability statement
The Veratrum oxysepalum data has been stored in nucleotide database of National Center of Biotechnology Information. GenBank accession number is MW147219. The SRA number is SRR14227219. All the information can be found on the website (https://www.ncbi.nlm.nih.gov/nuccore/).
Additional information
Funding
References
- Christopher MC, McDougal OM. 2014. Medicinal history of North American Veratrum. Phytochem Rev. 13(3):671–694.
- Cong Y, Wu Y, Shen S, Liu XP, Guo JG. 2020. A structure–activity relationship between the Veratrum alkaloids on the antihypertension and DNA damage activity in mice. Chem Biodiversity. 17(2):e1900473.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Editorial Committee of Chinese Academy of Sciences. 1980. Flora of China. Vol. 14. Beijing: Beijing Scientific Press.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20(17):3252–3255.
- Zomlefer WB, Whitten WM, Williams NH, Judd WS. 2003. An overview of Veratrum s.l. (Liliales: Melanthiaceae) and an infrageneric phylogeny based on ITS sequence Data. Syst Bot. 28(2):250–269.