Abstract
The parasitic wasp Anisopteromalus calandrae is a natural enemy of numbers store product pests. The mitochondrial genome (mitogenome) of A. calandrae was obtained by second-generation sequencing. The assembled mitogenome of A. calandrae is 15,954 bp long (GenBank accession: MW817149) and contains 37 typical animal mitochondrial genes. The order of the mitochondrial genes is identical to that of another species of Chalcidoidae (Pteromalus puparum). All protein-coding genes start with ATN codons, and end with TAA, except NAD4 and NAD5 with T.
The Anisopteromalus calandrae (Howard) is a well-known natural enemy of numbers important coleopterous stored product pests (Schöller et al. Citation2006), by parasitizing their larval or pupal stages. The cosmopolitan parasitic wasp has been investigated in biological control (Schöller et al. Citation2006), life-history traits (Narongplian et al. Citation2011), and other topics (Bressac et al., 2009). Data obtained from molecular fragments deepen the understanding of classification (Baur et al. Citation2014). This mitochondrial genome would supply more information on understanding its special character.
Samples of A. calandrae were originally collected alive from the culture flask of Sarocladium oryzae in National Engineering Laboratory for Rice and By-product Deep Processing, Central South University of Forestry and Technology, Changsha, Hunan province, China (28.14° N, 112.20° E). Samples are deposited at the Grain Insect specimens Museum (NEL6-19), Central South University of Forestry and Technology, China.
Whole genomic DNA was extracted with the CTAB method. The qualified DNA was sequenced using Illumina Novaseq. The clean reads were assembled using SPAdes (Version 3.13.1) (Bankevich et al. Citation2012). MITOS was performed to annotate the gene structure with five Invertebrates and other default parameters (Bernt et al. Citation2013).
The mitochondrial genome of A. calandrae consists of 15,954 bp in length with 82.9% of A + T (accession number MW817149), containing 13 PCGs (ATP6, ATP8, COXI-III, NAD1-6, NAD4L, and CYTB), two rRNAs genes (srRNA and lrRNA), and interspersed 22 tRNAs genes. The order of the mitochondrial genes is identical to that of another species of Chalcidoidae (Pteromalus puparum), including “cox1-trnL2-cox2-trnK-trnD-atp8-atp6-cox3” and “trnA-rrnL-trnL1-nad1” block shared in chalcidoid (Shimada and Fujii Citation1985; Yan et al. Citation2019). Furthermore, a non-coding region was found inserted between “trnA-rrnL” and “trnA2-CYTB.”
All of the 13 PCGs start with a typical ATN codon: six with ATT (NAD2, NAD3, ATP8, NAD5, NAD4l, and AND6), five with ATG (COX3, ATP6, COX1, NAD4, and COB), and two with ATA (COX2 and NAD1). Eleven PCGs stop with the complete termination codons TAA. The two exceptions are NAD4 and NAD5, which use T as a stop codon.
The second structure of 20 tRNA was predicted as typical cloverleaf secondary structures, except TrnS1 (AGA), which lacked a D ring and dihydrouridine (DHU) arm.
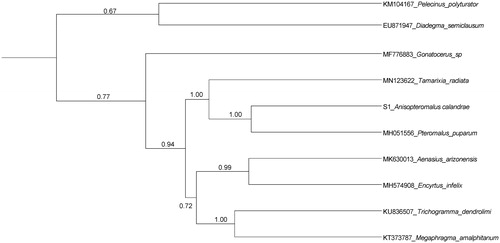
The phylogenetic relationships were reconstructed using Maximum Likelihood (ML) with MEGA7 (Kumar et al. Citation2016) and Bayesian inference (BI) with BEAST V 1.8.2 (Drummond et al. Citation2012) based on 13 PCGs dataset with nine Chalcidoidea from GenBank. The ML analyses were carried with the NJ model and 1000 bootstrap replications. Each BEAST analysis performed a lognormal relaxed clock model with 100 million generations (Gernhard Citation2008). The ML and BI analyses resulted in very similar topologies, and BI is better supported the relationships (). Both BI and ML tree strongly supported the sister relationship between A. calandrae and P. puparum (PP = 1.00, BS = 100), then sister to Tamarixia radiata (PP = 1.00, BS = 0.64). The clade of the three wasps is a well-supported sister to a clade comprising of Aenasius arizonensis and Encyrtus infelix, Trichogramma dendrolimi, and Megaphragma amalphitanum in BEAST tree (PP = 0.94, ). The other nodes were weakly supported, and more taxon sequences would be necessary.
Acknowledgments
We thank the anonymous reviewers for providing his/her valuable comments on the manuscript.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Data availability statement
The data that support the findings of this study are openly available in “NCBI” at https://www.ncbi.nlm.nih.gov/, reference accession number MW817149.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Baur H, Kranz-Baltensperger Y, Cruaud A, Rasplus JY, Timokhov AV, Gokhman VE. 2014. Morphometric analysis and taxonomic revision of Anisopteromalus Ruschka (Hymenoptera: Chalcidoidea: Pteromalidae) – an integrative approach. Syst Entomol. 39(4):691–709.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Drummond AJ, Suchard MA, Xie D, Rambaut A. 2012. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol. 29(8):1969–1973.
- Gernhard T. 2008. The conditioned reconstructed process. J Theor Biol. 253(4):769–778.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Narongplian N, Visarathanonth P, Amornsak W. 2011. Life cycle of Anisopteromalus calandrae (Howard) (Hymenoptera: Pteromalidae) on the maize weevil, Sitophilus zeamais Motschulsky (Coleoptera: Curculionidae). Bangkok: Kasetsart University. p. 232–238.
- Schöller ME, Flinn PW, Grieshop MJ, Žd'árková E. 2006. Chapter 9 – biological control of stored-product pests. In: Heaps JW, editor. Insect management for food storage and processing. 2nd ed. AACC International Press. 67–87.
- Shimada M, Fujii K. 1985. Niche modification and stability of competitive systems. I. Niche modification process. Res Popul Ecol. 27(1):185–201.
- Yan Z, Fang Q, Tian Y, Wang F, Chen X, Werren JH, Ye G. 2019. Mitochondrial DNA and their nuclear copies in the parasitic wasp Pteromalus puparum: a comparative analysis in Chalcidoidea. Int J Biol Macromol. 121:572–579.