Abstract
Notostomus gibbosus is a deep-sea shrimp, belonging to Caridea, Acanthephyridae. The whole complete mitochondrial genome of N. gibbosus was 17,956 bp in length, with 37 genes, containing 13 protein-coding genes, 22 tRNAs, and 2 rRNAs. The GC content of N. gibbosus was 39.43%. The genomic structure and gene arrangement were identical to those of Caridea species. The phylogenetic analysis of 13 protein-coding genes showed a close relationship to the genera Acanthephyra.
Notostomus gibbosus A. Milne-Edwards, 1881 (Caridea: Acanthephyridae) is a typical deep-sea shrimp ubiquitous in the global ocean (De Grave Citation2011; WoRMS Citation2020). This species is famous for its positive buoyancy, using ion replacement to balance body density (Sanders and Childress Citation1988). Notostomus gibbosus has a red soft integument, no cuticular photophores, and small eyes (Chan et al. Citation2010; Wong et al. Citation2015). The strengthened carapace and half-serrated mandible of N. gibbosus are well adapted to feeding on gelatinous organisms, such as deep-sea fishes (Lunina et al. Citation2020). Notostomus gibbosus contribute significantly to the bathypelagic food web and ecosystem function because of its widespread distribution and large individual biomass (Burghart et al. Citation2010). More than 52 complete mitochondrial genomes of Caridea have been determined till now (Sun et al. Citation2020). However, it remains unknown in the genus Notostomus. In this study, we reported the first complete mitochondrial genome of N. gibbosus and analyzed its phylogenetic relationship among Caridea.
A specimen of N. gibbosus was collected in January 2019 from the Indian Ocean (16.96°S, 89.69°E). The voucher specimen was stored in Key Laboratory of Science and Engineering for Marine Ecology and Environment, First Institute of Oceanography, MNR (Yuan Chao, [email protected], under the voucher number FIO-ECH-DY52DQSP02). Genomic DNA was extracted from the muscles using a QIAamp Fast DNA Tissue kit (Qiagen, Germany) according to the manufacturer’s instructions. A genomic library was then constructed. The mitochondrial genome of N. gibbosus was sequenced on the Illumina HiSeq 2500 sequencing platform (Illumina, USA) by Novogene Corporation (Beijing, China). The clean data were then assembled using the NOVOPlasty (Dierckxsens et al. Citation2017). This mitochondrial genome was annotated by MITOS2 and GeSeq (Tillich et al. Citation2017; Donath et al. Citation2019). The information of this mitochondrial genome was submitted to Genbank and the accession number is MW479468.
The circular mitochondrial genome of N. gibbosus was 17,956 bp in length with 39.43% GC content and contained 37 genes, including 13 protein-coding genes, 2 ribosomal RNA genes, and 22 transfer RNA genes. The composition and arrangement of genes were identical to those of other deep-sea Caridea shrimps. Our 16S rDNA and/or COI sequence were more than 99.5% identical to those of the partial sequence of N. gibbosus collected from the Gulf of Mexico (536 bp of 16S rDNA, Genbank accession number of MF197201 and 660 bp of COI, Genbank accession number of MH572548), and Taiwan waters (771 bp of 16S rDNA, Genbank accession number of GQ131894) (Chan et al. Citation2010).
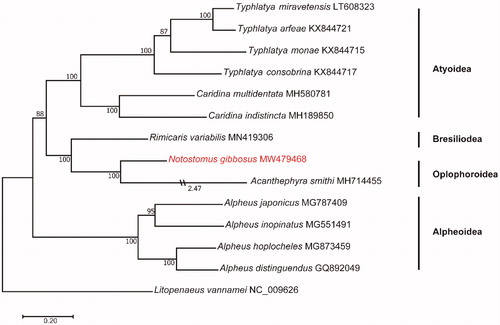
Phylogenetic relationships between N. gibbosus and 12 other Caridea shrimps, as well as one outgroup from the genus Litopenaeus, were analyzed using the maximum-likelihood method by IQTREE (Nguyen et al. Citation2015). Although data from Caridea mitochondrial genome were lacking, phylogenetic analysis established by 13 protein-coding genes resolved a close relationship of N. gibbosus to the genera Acanthephyra, together which formed a distinct group to the genera Rimicaris, then to the genera Typhlatya and Caridina in superfamily Atyoidae (). This study provided important information for further evolutionary and phylogenetic study of Notostomus.
Figure 1. The maximum-likelihood phylogenetic tree based on 13 protein-coding mitochondrial genes in 13 species in Caridea and one Litopenaeus mitogenome as an outgroup. The numbers at each branch represent bootstrap values (1000 replicates) and Genbank accession numbers for published sequences are incorporated. The branch lengths are proportional unless stated (for Acanthephyra smithi).
Disclosure statement
The authors report no conflict of interest. The authors alone are responsible for the content and writing of the paper.
Data availability statement
The raw data that support this study are openly available in SRA of NCBI under accession no. PRJNA728383. The genome sequence data are deposited in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/nuccore/ under the accession no. MW479468.
Additional information
Funding
References
- Burghart SE, Hopkins TL, Torres JJ. 2010. Partitioning of food resources in bathypelagic micronekton in the eastern Gulf of Mexico. Mar Ecol Prog Ser. 399:131–140.
- Chan T-Y, Lei HC, Li CP, Chu KH. 2010. Phylogenetic analysis using rDNA reveals polyphyly of Oplophoridae (Decapoda:Caridea). Invert Systematics. 24(2):172–181.
- De Grave S. 2011. Carideorum catalogus: the recent species of the dendrobranchiate, stenopodidean, procarididean and caridean shrimps (Crustacea: Decapoda). Zoologische Mededelingen. 85:195–589.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Donath A, Jühling F, Al-Arab M, Bernhart SH, Reinhardt F, Stadler PF, Middendorf M, Bernt M. 2019. Improved annotation of protein-coding genes boundaries in metazoan mitochondrial genomes. Nucleic Acids Res. 47(20):10543–10552.
- Lunina AA, Kulagin DN, Vereshchaka AL. 2020. Phylogenetic revision of the shrimp genera Ephyrina, Meningodora and Notostomus (Acanthephyridae: Caridea). Zoolog J Linnean Soc.zlaa161.
- Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Sanders NK, Childress JJ. 1988. Ion replacement as a buoyancy mechanism in a pelagic deep-sea crustacean. J Experimental Biol. 138(1):333–343.
- Sun SE, Cheng J, Sun S, Sha Z. 2020. Complete mitochondrial genomes of two deep-sea pandalid shrimps, Heterocarpus ensifer and Bitias brevis: insights into the phylogenetic position of Pandalidae (Decapoda: Caridea). J Ocean Limnol. 38(3):816–825.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq - versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
- Wong JM, Pérez-Moreno JL, Chan TY, Frank TM, Bracken-Grissom HD. 2015. Phylogenetic and transcriptomic analyses reveal the evolution of bioluminescence and light detection in marine deep-sea shrimps of the family Oplophoridae (Crustacea: Decapoda). Mol Phylogenet Evol. 83:278–292.
- WoRMS. 2020. Notostomus gibbosus A. Milne-Edwards, 1881; [Accessed 2020 December 24]. http://www.marinespecies.org/aphia.php?p=taxdetails&id=212899
