Abstract
Erionota torus (Evans, 1941) is a banana pest and is mainly distributed in Southeast Asia and the Pacific regions. The complete mitogenome of E. torus (GenBank accession number MW586888) is 15,987 bp in size, including 13 protein-coding genes, 22 transfer RNAs, 2 ribosomal RNAs genes, and a noncoding A + T-rich region. The A + T-rich region is located between 12S rRNA and tRNAMet. The base composition of the whole E. torus mitogenome is 39.68% for A, 7.30% for G, 41.55% for T, and 11.47% for C, with a high AT content of 81.23%. The phylogeny analysis indicated that E. torus had a close relationship with Notocrypta curvifascia. The present data could contribute to the further detailed phylogeographic analysis and provide a comprehensive control strategy for this banana pest.
Banana (Musa spp.) is an ideal and low-cost crop and accounts for the fourth most important food in the world today (Tripathi et al. Citation2021). Especially in developing countries, many populations depend mostly on bananas as food and feed source (Mohapatra et al. Citation2010). The banana skipper or banana leaf-roller, Erionota torus (Evans, 1941) (Lepidoptera: Hesperiidae), is a common banana pest in Southeast Asia and Pacific regions, ranging from Papua New Guinea and Australia, Sikkim to south China, Myanmar, Malaysia, and Vietnam (Inoue and Kawazoe Citation1970; Okolle et al. Citation2006; Corbet and Pendlebury Citation1992). Almost all banana cultivars could be attacked by this skipper (Sivakumar et al. Citation2014). The larvae of E. torus cause considerable damage to banana foliage by rolling the leaf while feeding on it (Chiang and Hwang Citation1991) and could lead to yield loss of about 20% (Okolle et al. Citation2010). Elucidating the sequence and structure of E. torus mitogenome is important for the diversity and phylogeographic analysis of this banana pest, thus providing information for its comprehensive control.
The specimen of E. torus in the present work was obtained from Honghe, Yunnan, China (N 22°77′, E 103°24′), and deposited in the insect museum (handled by Jin-Hua Zhang, email: [email protected]) in Agricultural Environment and Resources Institute, Yunnan Academy of Agricultural Sciences, with a voucher number AERI-G-20200518. Sequencing work of the complete mitogenome of E. torus was performed by Illumina Nextseq500 in Beijing Microread Genetics Co., Ltd., with a total data volume of 4 G (150 bp Reads). High-quality reads were assembled from scratch using IDBA-UD and SPAdes (Gurevich et al. Citation2013). Protein-coding genes (PCGs) of the E. torus mitogenome were identified using BLAST search in NCBI, and tRNA genes were identified using the tRNAscan-SE search server (Schattner et al. Citation2005). The final assembled mitogenome was also verified on the MITOS web server (Bernt et al. Citation2013).
The E. torus mitogenome is 15,987 bp in size (GenBank accession number MW586888), including 13 typical invertebrate PCGs, 22 transfer RNA genes, 2 ribosomal RNA genes, and a noncoding control region (A + T-rich). The A + T content of the whole E. torus mitogenome is 81.23%, showing an obvious AT mutation bias (Eyre-Walker Citation1997). The A + T-rich region exhibits the highest A + T content (94.89%) in the E. torus mitogenome.
All the 13 PCGs use standard ‘ATN’ as start codons and have the common mitochondrial stop codon ‘TAA.’ All the tRNAs except tRNASer (GCU) could be folded into the typical cloverleaf secondary structures. The unusual tRNASer (GCU) had completely lost the dihydrouridine (DHU) stem and loop. The 12S rRNA gene is located between tRNAVal and the A + T-rich region, while the 16S rRNA is located between tRNAVal and tRNALeul. The locations of these two rRNA genes in E. torus mitogenome are quite different from the ancestral insect’s mitogenome (Boore Citation1999).
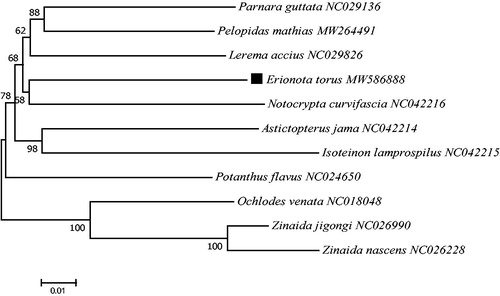
Based on the concatenated 13 mitochondrial PCGs sequences of 11 species from Hesperiinae, the neighbor-joining method was used to construct the phylogenetic relationship between E. torus and 10 other Hesperiinae species (). The phylogenetic analysis was performed by MEGA7 software (Kumar et al. Citation2016). The potential saturation of any PCG was assessed using DAMBE5 software (Xia Citation2013). The phylogeny analysis indicated that E. torus had a close relationship with Notocrypta curvifascia (), which adds new information to the evolutionary lineage research of E. torus (Jaleel et al. Citation2019). This mitogenome data might be also valuable for further phylogeography analyses and provide a comprehensive control strategy for this banana pest.
Figure 1. Phylogenetic tree showing the relationship between E. torus and 10 other hesperiinae species based on neighbor-joining method performed using 500 bootstrap replicates. Ochlodes venata, Zinaida jigongi and Zinaida nascens were used as outgroups. GenBank accession numbers of each sequence were listed in the tree behind their corresponding species names.
Disclosure statement
No potential conflict of interest was reported by the author(s). The views expressed in this document cannot be taken to reflect the official opinions of the funding organizations.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov under the accession no. MW586888. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA721010, SRR14202297, and SAMN18695551 respectively. CRPs are implemented with support from the CGIAR Trust Fund and through bilateral funding agreements. For details, please visit https://ccafs.cgiar.org/donors.
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Boore JL. 1999. Animal mitochondrial genomes. Nucleic Acids Res. 27(8):1767–1780.
- Chiang HS, Hwang MT. 1991. The banana skipper, Erionota torus Evans (Hesperidae: Lepidoptera): establishment, distribution and extent of damage in Taiwan. Pans Pest Articles News Summ. 37(3):207–210.
- Corbet AS, Pendlebury HM. 1992. The butterflies of the Malay peninsula. 4th ed. Kuala Lumpur (Malaysia): Malayan Nature Society.
- Eyre-Walker A. 1997. Differentiating between selection and mutation bias. Genetics. 147(4):1983–1987.
- Gurevich A, Saveliev V, Vyahhi N, Tesler G. 2013. QUAST: quality assessment tool for genome assemblies. Bioinformatics. 29(8):1072–1075.
- Inoue S, Kawazoe A. 1970. Hesperiid butterflies from South Vietnam. Trans Lep Soc Jap. 21:1–14.
- Jaleel KA, Ghosh SM, Joseph VJ. 2019. DNA barcoding and evolutionary lineage of banana skipper Erionota torus (Evans) (Lepidoptera: Hesperiidae) from Malabar, a part of Southern Western Ghats, India. Int J Sci Res Biol Sci. 6(4):29–32.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Mohapatra D, Mishra S, Sutar N. 2010. Banana and its by-product utilisation: an overview. J Sci Ind Res India. 69:323–329.
- Okolle JN, Mansor M, Ahmad AH. 2006. Spatial distribution of banana skipper Erionota thrax L. (Lepidoptera: Hesperiidae) and its parasitoids in a Cavendish banana plantation, Penang, Malaysia. Insect Sci. 13(5):381–389.
- Okolle JN, Ahmad AH, Mansor M, Tripathi L. 2010. Bioecology and management of the banana skipper (Erionota thrax). Tree Forestry Sci Biotechnol. 4(1):22–31.
- Schattner P, Brooks AN, Lowe TM. 2005. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 33:W686–W689.
- Sivakumar T, Jiji T, Anitha N. 2014. Field observations on banana skipper Erionota thrax L. (Hesperiidae: Lepidoptera) and its avian predators from southern peninsular India. Curr Biotica. 3(8):220–227.
- Tripathi L, Ntui VO, Tripathi JN, Kumar PL. 2021. Application of CRISPR/Cas for diagnosis and management of viral diseases of banana. Front Microbiol. 27(11):609784.
- Xia X. 2013. DAMBE5: a comprehensive software package for data analysis in molecular biology and evolution. Mol Biol Evol. 30(7):1720–1728.
