Abstract
The complete mitochondrial genome of Taxoblenus sinicus Wei & Nie, Citation1999 was described. The circular genome is 15,878 bp with an A + T content of 80.4%. It contains 37 genes, and an 859 bp control region. The trnI (+)-trnQ (−)-trnM (+) cluster rearranges to trnM (+)-trnQ (−)-CR-trnI (+). Phylogenetic analysis demonstrates that Allantinae is a sister group of Tenthredininae and T. sinicus is one of the basal lineages of Allantinae.
Taxoblenus Wei & Nie, Citation1999 (Wei and Nie Citation1999) is a member of the subtribe Taxonina of the tribe Allantini of Allantinae. Species of Taxoblenus occur mostly in the alpine meadow region of Western China. The mitochondrial genome of Taxoblenus would be helpful to analyze the phylogeny of Allantinae. The complete mitochondrial genome of Taxoblenus sinicus Wei & Nie, Citation1999 was reported herein.
Samples of T. sinicus (CSCS-Hym-MC0193) were collected in Sigou, Zhugu, Menyuan, Qinghai, China (37.11 N 102.35 E) in July 2019. The specimen was deposited at the Asia Sawfly Museum, Nanchang (ASMN) (Meicai Wei, [email protected]) under the voucher number CSCS-Hym-MC0193. Whole genomic DNA was extracted from the thorax muscle of a female adult using the DNeasyR Blood &Tissue Kits (Qiagen, Valencia, CA). Genomic DNA was prepared in 150 bp paired-end libraries, tagged, and analyzed with the high-throughput Illumina Hiseq 4000 platform. A total of 4,734,909 raw reads (SRR14085847) were obtained. DNA sequences were assembled using MitoZ (Meng et al. Citation2019) and verified by Geneious Prime 2019.2.1 (https://www.geneious.com). Annotations of tRNAs were generated in MITOS web server (Bernt et al. Citation2013) and revised where necessary. The eleven unsaturated nucleotide sequences (atp6, cob, cox1, cox2, cox3, nad1, nad2, nad3, nad4, nad4l, and nad5) of 37 species were aligned using the MEGA7.0.26 (Kumar et al. Citation2016). The phylogenetic tree was constructed using RAxML (Stamatakis Citation2014) under the model GTR, and MrBayes (Ronquist et al. Citation2012).
The sequence yield by MitoZ was 15,325 bp and contained 37 genes with a partial control region (CR). The obtained sequences were thoroughly examined by reassembly using Allantus togatus (MW464859) and Ferna bicoloricornis (unpublished) as reference sequences (coverage were 6644 and 34,300, respectively). Reassembly using trnI and trnQ as references extended contigs and obtained a 126 bp overlap. After manual verification, we obtained a control region of 859 bp in length. To verify the reliability of the results, we used the control region itself as a reference, and received a high-quality mapping with the flanking tRNAs were found.
The circular genome is 15,878 bp, including 37 genes and a control region. The gene rearrangement only occurred in tRNA cluster as trnM (+)-trnQ (−)-CR-trnI (+). Except for Asiemphytus rufocephalus (incomplete), there are two existing gene rearrangement patterns in Allantinae. Due to the high diversity of Tenthredininae, the current sample coverage may not be sufficient to speculate the systematic significance of gene order patten. The A + T content of the whole mitochondrial genome is 80.40% (42.70% A, 11.80% C, 7.90% G, and 37.7% T), indicating significant A + T bias. Four start codons for protein-coding genes (PCGs) are used, ATG (atp6, cob, cox3, nad2, and nad4); ATA (cox2, nad1, nad4l, nad5, and nad6); ATT (atp8 and nad3) and TTG (cox3). All PCGs use TAA as a stop codon except for nad4 (T).
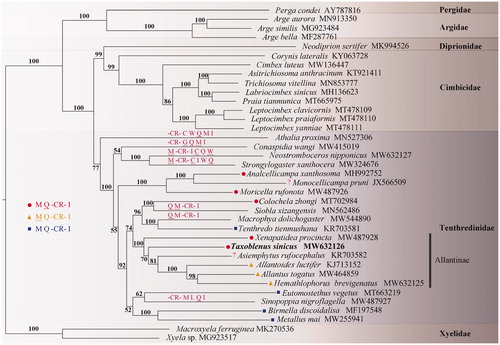
Phylogenetic tree based on mitochondrial genome sequences of 37 Tenthredinoidea species with two Xyelidae species as outgroup. The maximum-likelihood (ML) and Bayesian methods (BI) for phylogenetic analysis resulted in trees with similar topologies, and only the ML tree is shown in . All conduction supports that (1) Allantinae is a sister group of Tenthredininae; (2) T. sinicus is one of the basal lineages of Allantinae (Wei and Nie Citation1998).
Figure 1. A maximum-likelihood (ML) tree based on the combined data of eleven unsaturated nucleotides. Numbers at the left of nodes are bootstrap support value. Partial gene orders are marked on the branches either directly or using the corresponding icons. The accession number for each species is indicated after the Latin name.
Disclosure statement
No potential conflict of interest was reported by the authors. The authors alone are responsible for the content and writing of the article.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov under the accession number MW632126. The associated BioProject, SRA, and BioSample numbers are PRJNA714511, SRR14085847, and SAMN18318555, respectively. All related files had been uploaded to Science Data Bank (http://www.scidb.cn/s/pMvqE7v).
Additional information
Funding
References
- Bernt M, Donath A, Juhling F, Externbrink F, Florentz C, Fritzsch G, Putz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Meng G, Li Y, Yang C, Liu S. 2019. MitoZ: a toolkit for animal mitochondrial genome assembly, annotation and visualization. Nucleic Acids Res. 47(11):e63.
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Hohna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Wei M, Nie H. 1998. Generic list of Tenthredinoidea in new systematic arrangement with synonyms and distribution data. J Cent South Univ. 18(3):23–31.
- Wei M, Nie H. 1999. A new genus and seven new species of Allantinae (Hymenoptera: Tenthredinidae) from China. J Cent South Univ. 19(3):9–14.
