Abstract
The complete mitogenome sequence of the lesser bandicoot rat (Bandicota bengalensis Gray and Hardwicke, 1833) was determined using long PCR. The genome was 16,327 bp in length and contained 13 protein-coding genes, 2 ribosomal RNA genes, 22 transfer RNA genes, 1 origin of L strand replication and 1 control region. The overall base composition of the heavy strand is A (34.2%), C (24.9%), T (28.5%) and G (12.4%). The base compositions present clearly the A–T skew, which is most obviously in the control region and protein-coding genes. Mitochondrial genome analyses based on MP, ML, NJ and Bayesian analyses yielded identical phylogenetic trees. This study verifies the evolutionary status of Bandicota bengalensis in Muridae at the molecular level. The mitochondrial genome would be a significant supplement for the Bandicota bengalensis genetic background. The two Bandicota species formed a monophyletic group with the high bootstrap value (100%) in all examinations.
The lesser bandicoot rat (Bandicota bengalensis Gray and Hardwicke, 1833) is a murid rodent distributed mostly in Asia that can cause substantial negative economic impact in urban and rural areas. In this paper, A muscle sample was obtained from a female Bandicota bengalensis captured from Ruili regions in Yunnan Province, China (24°01′12″ N, 97°85′18″ E). The muscle tissue was preserved in 95% ethanol and stored at −75 °C before use. The specimen and its DNA is stored in Animal and Plant Herbarium of Mudanjiang Normal University. The voucher number is XBCS2019004 (liuzhu, [email protected]). Genomic DNA was extracted from muscle using the EasyPure genomic DNA kit (TransGen Biotech Co., Beijing, China). The mitogenomes were sequencing by Illumina NovaSeq 6000 platform (Ruiboxingke Biotechnology Co. Ltd., Beijing, China) using a primer walking strategy and the long and accurate PCR. The draft sequence was manually corrected. The complete mitochondrial genome sequence was annotated using Sequin.
The mitochondrial genome is a circular double-stranded DNA sequence that is 16,327 bp long including 13 protein-coding genes, 2 rRNA genes, 22 tRNA genes, 1 origin of L strand replication and 1 control region. The accurate annotated mitochondrial genome sequence was submitted to GenBank with accession number MW364625. The arrangement of the multiple genes is in line with other Muridae species (Robins et al. Citation2008; Chen et al. Citation2012; Jing et al. Citation2015; Chang et al. Citation2016; Yong et al. Citation2016; Zhang et al. Citation2016; Wei et al. Citation2017; Lv et al. Citation2019) and most mammals (Mouchaty et al. Citation2000; Nikaido et al. Citation2001; Nikaido et al. Citation2003; Fontanillas et al. Citation2005; Cabria et al. Citation2006; Meganathan et al. Citation2012; Yoon et al. Citation2013; Xu et al. Citation2012, Citation2013; Kim et al. Citation2013, Citation2017; Hou et al. Citation2016; Huang et al. Citation2014, Citation2016; Xu et al. Citation2016; Liu et al. Citation2016; Liu, Tian, Jin, Jin, et al. Citation2017; Liu, Tian, Jin, Dong, et al. 2017; Liu, Wang, et al. Citation2017; Liu et al. Citation2018; Liu, Dang, et al. Citation2019; Liu, Qin, et al. Citation2019; Jin et al. Citation2017; Gutiérrez et al. Citation2018; Jia et al. Citation2018).
The control region of Bandicota bengalensis mitochondrial genome was located between the tRNA-Pro and tRNA-Phe genes, and contains only promoters and regulatory sequences for replication and transcription, but no structural genes. Three domains were defined in Bandicota bengalensis mitochondrial genome control region (Zhang et al. Citation2009): the extended termination-associated sequence (ETAS) domain, the central conserved domain (CD) and the conserved sequence block (CSB) domain.
The total length of the protein-coding gene sequences was 11,405 bp. Most protein-coding genes initiate with ATG except for ND1, ND3 and ND5, which began with GTG or ATT. Eleven protein-coding genes terminated with TAA. The incomplete stop codons (T– –) were used in COX3, ND4 and Cyt b. A strong bias against A at the third codon position was observed in the protein-coding genes. The frequencies of CTA (Leu), ATT (Ile), TTA (Leu) and ATA (Met) were higher than those of other codons. The length of tRNA genes varied from 58 to 77 bp.
Most Bandicota bengalensis mitochondrial genes were encoded on the H strand, except for the ND6 gene and eight tRNA genes, which were encoded on the L strand. Some reading frame intervals and overlaps were found. One of the most typical was between ATP8 and ATP6. The L-strand replication origin (OL) was 32 bp long and had the potential to fold into a stable stem-loop secondary structure. The total base composition of Bandicota bengalensis mitochondrial genome was A (34.2%), C (24.9%), T (28.5%) and G (12.4%). The base compositions clearly present the A-T skew, which was most obviously in the control region and protein coding genes.
In order to explore the evolution of Muridae species, especially the evolution of genus Bandicota from China, here, we investigate the molecular phylogenetics of Chinese Bandicota bengalensis using complete mitochondrial genome sequence of 36 species. All sequences generated in this study have been deposited in the GenBank ().
Figure 1. Phylogenetic tree generated using the Maximum Parsimony method based on complete mitochondrial genomes. The out group is Ochotona coreana (MT017929).
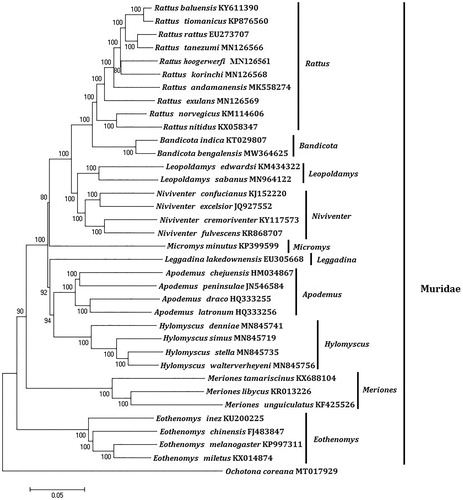
Mitochondrial genome analyses based on MP, ML, NJ and Bayesian analyses yielded identical phylogenetic trees, indicating a close phylogenetic affinity of species. The phylogram obtained from ML method is shown in . It shows that one major phyletic lineages were present in Muridae. In this study, the 10 genera (Rattus, Niviventer, Bandicota, Hylomyscus, Leopoldamys, Apodemus, Micromys, Eothenomys, Leggadina, and Meriones) included in Muridae form independent branches. Bandicota comprised Bandicota bengalensis and Bandicota indica was supported by bootstrap values of 100%. This study verifies the evolutionary status of Bandicota bengalensis in Muridae at the molecular level. The mitochondrial genome would be a significant supplement for the Bandicota bengalensis genetic background. The two Bandicota species formed a monophyletic group with the high bootstrap value (100%) in all examinations.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Data availability statement
The data that support the findings of this study are openly available in GenBank at https://www.ncbi.nlm.nih.gov/, reference number MW364625.
Additional information
Funding
References
- Cabria MT, Rubines J, Gómez-Moliner B, Zardoya R. 2006. On the phylogenetic position of a rare Iberian endemic mammal, the Pyrenean desman (Galemys pyrenaicus). Gene. 375:1–13.
- Chang P, Li J, Hwang D. 2016. The complete mitochondrial genome of western Mediterranean mouse, Mus spretus (Rodentia: Muridae). Mitochondrial DNA Part A. 27(3):2135–2136.
- Chen W, Sun Z, Liu Y, Yue B, Liu S. 2012. The complete mitochondrial genome of the large white-bellied rat, Niviventer excelsior (Rodentia: Muridae). Mitochondrial DNA. 23(5):363–365.
- Fontanillas P, Depraz A, Giorgi MS, Perrin N. 2005. Nonshivering thermogenesis capacity associated to mitochondrial DNA haplotypes and gender in the greater white-toothed shrew, Crocidura russula. Mol Ecol. 14(2):661–670.
- Gutiérrez J, Lamelas L, Aleix-Mata G, Arroyo M, Marchal JA, Palomeque T, Lorite P, Sánchez A. 2018. Complete mitochondrial genome of the Iberian Mole Talpa occidentalis (Talpidae, Insectivora) and comparison with Talpa europaea. Genetica. 146(4–5):415–423.
- Hou Q, Tu F, Liu Y, Liu S. 2016. Characterization of the mitogenome of Uropsilus gracilis and species delimitation. Mitochondrial DNA Part A. 27(3):1836–1837.
- Huang T, Dang X, An M, Chen L, Zhang J. 2016. The complete mitochondrial genome of the Sorex araneus. Mitochondrial DNA A DNA Mapp Seq Anal. 27(5):3655–3656.
- Huang T, Yan CC, Tan Z, Tu FY, Yue BS, Zhang XY. 2014. Complete mitochondrial genome sequence of Nectogale elegans. Mitochondrial DNA. 25(4):253–254.
- Jia X, Yang L, Shi H. 2018. The complete mitochondrial genome of Anderson’ s shrew mole, Uropsilus andersoni (Talpidae). Conservation Genet Resour. 10(3):583–585.
- Jin ZM, Liu Z, Ma JZ. 2017. Sequencing and analysis of the complete mitochondrial genome of the masked shrew (Sorex caecutiens) from China. Mitochondrial DNA B Resour. 2(2):486–488.
- Jing J, Song X, Yan C, Lu T, Zhang X, Yue B. 2015. Phylogenetic analyses of the harvest mouse, Micromys minutus (Rodentia: Muridae) based on the complete mitogenome sequences. Biochem Syst Ecol. 62:121–127.
- Kim TW, Kim YK, Oh DJ, Park JH, Kim D, Adhikari P, Kim G, Park SM, Lee JW, Jung YH, et al. 2017. Complete mitochondrial genome of the Ussuri white-toothed shrew Crocidura lasiura (Insectivora, Soricidae). Mitochondrial DNA A DNA Mapp Seq Anal. 28(2):216–217.
- Kim HR, Park JK, Cho JY, Chul Park Y. 2013. Complete mitochondrial genome of an Asian Lesser White-toothed Shrew, Crocidura shantungensis (Soricidae). Mitochondrial DNA. 24(3):202–204.
- Liu Z, Bai W, Wang AN, Tian XM, Li DW. 2018. Sequencing and analysis of the complete mitochondrial genome of the taiga shrew (Sorex isodon) from China. Mitochondrial DNA Part B. 3(1):466–468.
- Liu Z, Dang YQ, Li JJ. 2019. Sequencing and analysis of the complete mitochondrial genome of the Eurasian least shrew (Sorex minutissimus) from China. Mitochondrial DNA Part B. 4(1):178–180.
- Liu SY, Jin W, Liao R, Sun ZY, Zeng T, Fu JR, Liu Y, Wang X, Li PF, Tang MK, et al. 2017. Phylogenetic study of Ochotona based on mitochondrial Cyt b and morphology with a description of one new subgenus and five new species. Acta Theriologica Sinica. 37(1):1–43.
- Liu Z, Qin KS, Li JJ, Dong M. 2019. Sequencing and analysis of the complete mitochondrial genome of the Siberian large-toothed shrew (Sorex daphaenodon) from China. Mitochondrial DNA Part B. 4(1):542–544.
- Liu Z, Tian XM, Jin ZM, Dong M, Zhang JS. 2017b. Sequencing and analysis of the complete mitochondrial genome of the Ussuri shrew (Sorex mirabilis) from China. Mitochondrial DNA Part B. 2(2):645–647.
- Liu Z, Tian XM, Jin JL, Jin ZM, Li DW, Zhang JS. 2017a. Sequencing and analysis of the complete mitochondrial genome of the slender shrew (Sorex gracillimus) from China. Mitochondrial DNA Part B. 2(2):642–644.
- Liu Z, Wang AN, Zhang JS, Yang X, Liu H. 2017. Sequencing and analysis of the complete mitochondrial genome of flat-skulled shrew (Sorex roboratus) from China. Mitochondrial DNA Part B. 2(1):369–371.
- Liu Z, Zhao W, Liu P, Li S, Xu C. 2016. The complete mitochondrial genome of Eurasian water shrew (Neomys fodiens). Mitochondrial DNA A DNA Mapp Seq Anal. 27(4):2381–2382.
- Lv X, Li H, Li Y, Wang J, Wang X, Li Y. 2019. Characterization of the complete mitochondrial genome of Indochinese Forest Rat, Rattus andamanensis (Rodentia: Muridae) and its phylogenetic analysis. Mitochondrial DNA Part B. 4(1):1455–1456.
- Meganathan PR, Pagan HJT, McCulloch ES, Stevens RD, Ray DA. 2012. Complete mitochondrial genome sequences of three bats species and whole genome mitochondrial analyses reveal patterns of codon bias and lend support to a basal split in Chiroptera. Gene. 492(1):121–129.
- Mouchaty SK, Gullberg A, Janke A, Arnason U. 2000. The phylogenetic position of the Talpidae within Eutheria based on analysis of complete mitochondrial sequences. Mol Biol Evol. 17(1):60–67.
- Nikaido M, Cao Y, Harada M, Okada N, Hasegawa M. 2003. Mitochondrial phylogeny of hedgehogs and monophyly of Eulipotyphla. Mol Phylogenet Evol. 28(2):276–284.
- Nikaido M, Kawai K, Cao Y, Harada M, Tomita S, Okada N, Hasegawa M. 2001. Maximum likelihood analysis of the complete mitochondrial genomes of eutherians and a reevaluation of the phylogeny of bats and insectivores. J Mol Evol. 53(4–5):508–506.
- Robins JH, McLenachan PA, Phillips MJ, Craig L, Ross HA, Matisoo-Smith E. 2008. Dating of divergences within the Rattus genus phylogeny using whole mitochondrial genomes. Mol Phylogenet Evol. 49(2):460–466.
- Wei H, Li F, Wang X, Wang Q, Chen G, Zong H, Chen S. 2017. The characterization of complete mitochondrial genome and phylogenetic relationship within Rattus genus (Rodentia: Muridae). Biochem Syst Ecol. 71:179–186.
- Xu Y, Huang X, Hu Y, Tu F. 2016. Description of the mitogenome of Gansu mole (Scapanulus oweni). Mitochondrial DNA Part A. 27(3):2083–2084.
- Xu CZ, Zhang HH, Ma JZ. 2013. The complete mitochondrial genome of Martes flavigula. Mitochondrial DNA. 24(3):240–242.
- Xu CZ, Zhang HH, Ma JZ, Liu ZH. 2012. The complete mitochondrial genome of sable, Martes zibellina. Mitochondrial DNA. 23(3):167–169.
- Yong B, Wei H, Jia Q, Chen S. 2016. Sequencing and analysis of complete mitochondrial genome of Niviventer fulvescens (Muridae). Mitochondrial DNA A DNA Mapp Seq Anal. 27(5):3650–3651.
- Yoon KB, Kim HR, Kim JY, Jeon SH, Park YC. 2013. The complete mitochondrial genome of the Ussurian tube-nosed bat Murina ussuriensis (Chiroptera: Vespertilionidae) in Korea. Mitochondrial DNA. 24(4):397–399.
- Zhang YZ, Lou JY, Zhang CM, Li JF, Yin HL, Wang YL. 2016. Complete mitochondrial genome sequence of the Rattus norvegicus SILN strain with central nervous system disorder. Mitochondrial DNA Part A. 27(3):1610–1611.
- Zhang HH, Xu CZ, Ma JZ. 2009. Structure of the mtDNA control region and phylogeny of the Mustelidae species. Acta Ecol Sin. 29:3585–3592.
