Abstract
The mitogenome of Meleonoma mirabilis was determined in this study. It was 15,268 bps long and strongly AT biased. It harbored 13 PCGs, 22 tRNAs, 2 rRNAs, and 1 non-coding control region (334 bps). BI phylogenetic analysis based on 13 concatenated PCGs from 24 moth species indicated that M. mirabilis was clustered in the family Autostichidae, which was consistent with the latest phylogenetic study.
Meleonoma mirabilis (Wang Citation2003), a small moth species, belongs to the genus Meleonoma Meyrick (Autostichidae, Lepidoptera), which had experienced substantial expansion in 2020. In that year, more than 100 species (most from China) were described as new or transferred from the other genus, and 8 species groups were set up in necessity to accommodate the rapid accumulation of new species, and to facilitate innergeneric taxonomy (e.g. Wang and Zhu Citation2020a, Citation2020b; Wang et al. Citation2020). Division of species groups was based on morphological analysis plus molecular evidence of one single cox1 locus (Wang et al. Citation2020). More molecular loci should be involved to fully resolve this complex genus. Nevertheless, no mitogenome of Meleonoma was available so far. In compensation, we present herein the mitogenome of M. mirabilis which is a common species in South China (Wang et al. Citation2020). The adults were collected from Leigong Mountain Nature Reserve (26°22′23″‘N 108°11′52″‘E 1530 m), Guizhou, China in 2020, using light trap. The specimens were then deposited in 99% ethanol under −20 °C in the Insect Collection of Guizhou University of Traditional Chinese Medicine, Guiyang, China (Aihui Yin, [email protected]) under the voucher number GZUTCM:M59-62.
The genome sequencing was performed at Sangon Biotech (Shanghai) Co., Ltd., China, on Illumina HiSeq2500 platform. The de novo assembly was carried out with SPAdes V.3.14.1 (Bankevich et al. Citation2012) and NOVOPlasty V.4.0 (Dierckxsens et al. Citation2017). Gap filling and correctness check were executed manually with the aid of BWA V.0.7.17 (Li Citation2013), samtools V.0.1.19 (Li et al. Citation2009) and Pilon V.1.23 (Walker et al. Citation2014). MITOS WebServer (http://mitos2.bioinf.uni-leipzig.de/index.py) was utilized for annotation.
The double stranded circular mitogenome of M. mirabilis (GenBank: MW366996), was 15,268 bps long, and strongly AT biased (AT 77.1%, CG only 22.9%). It harbored the typical set of metazoan genes (13 PCGs, 22 tRNAs and two rRNAs) (Wolstenholme Citation1992), and one non-coding A + T rich control region (334 bps, AT 93.4%). Most PCGs of M. mirabilis started at conventional ATN start codon, only cox1 initiated with CGA. All PCGs used the typical TAA or TAG stop codon at termination, except for cox1, cox2, nad4, and nad5, which terminated with an incomplete single T residue. Twenty-one out of 22 tRNAs were folded into the typical cloverleaf structure, leaving TrnS1 the only exception, as it lacked the DHU arm. Gene order TrnM-TrnI-TrnQ rather than the more ancestral non-Ditrysian trnI-trnQ-trnM was also recognized in M. mirabilis (Park et al. Citation2016).
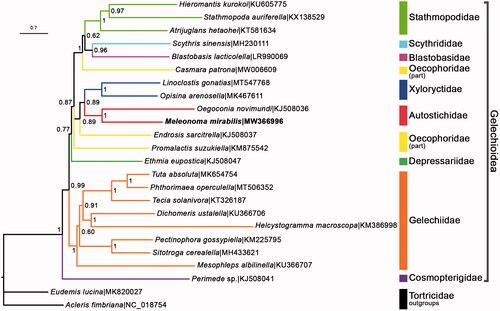
The Bayesian inference analysis was conducted via MrBayes V.3.2.7 (Ronquist et al. Citation2012) using ‘GTR + I + G’ substitution model within the superfamily Gelechioidea. Thirteen concatenated PCGs from M. mirabilis and other 23 species from GenBank were used to rebuild the phylogenetic tree (). Based on the currently available material, it showed that Autostichidae, Gelechiidae, Stathmopodidae, and Xyloryctidae which had multiple representatives were monophyletic. However, Oecophoridae was recovered as polyphyly. M. mirabilis was clustered in Autostichidae, which was consistent with the latest study (Wang and Li Citation2020).
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/nuccore/MW366996 under the accession No. MW366996. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA729565, SRR14517488, and SAMN19133288, respectively.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv. 13033997.
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup. 2009. The sequence alignment/map format and SAMtools. Bioinformatics. 25(16):2078–2079.
- Park JS, Kim MJ, Jeong SY, Kim SS, Kim I. 2016. Complete mitochondrial genomes of two gelechioids, Mesophleps albilinella and Dichomeris ustalella (Lepidoptera: Gelechiidae), with a description of gene rearrangement in Lepidoptera. Curr Genet. 62(4):809–826.
- Ronquist F, Teslenko M, Mark PV, Ayres DL, Darling A, Hohna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542.
- Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, Sakthikumar S, Cuomo CA, Zeng Q, Wortman J, Young SK, et al. 2014. Pilon: An integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLOS One. 9(11):e112963.
- Wang QY, Li HH. 2020. Phylogeny of the superfamily Gelechioidea (Lepidoptera: Obtectomera), with an exploratory application on geometric morphometrics. Zool Scr. 49(3):307–328.
- Wang SX. 2003. A study of Cryptolechia Zeller (Lepidoptera: Oecophoridae) in China (I), with descriptions of fifteen new species. Entomol Sin. 10(3):195–213.
- Wang SX, Zhu XJ. 2020a. Study of the genus Meleonoma Meyrick, 1914 (Lepidoptera: Autostichidae) from China, with descriptions of fifteen new species. Zootaxa. 4838(3):331–357.
- Wang SX, Zhu XJ. 2020b. Study of the genus Meleonoma Meyrick, 1914 (Lepidoptera: Autostichidae) from China, with descriptions of twenty-one new species (II). Zootaxa. 4881(2):257–289.
- Wang SX, Zhu XJ, Zhao BX, Yang XF. 2020. Taxonomic review of the genus Meleonoma Meyrick (Lepidoptera: Autostichidae), with a checklist of all the described species. Zootaxa. 4763(3):371–393.
- Wolstenholme DR. 1992. Animal mitochondrial DNA: structure and evolution. Int Rev Cytol. 141(6):173–216.