Abstract
The complete mitochondrial genome of Nais communis was analyzed using the Illumina Hiseq 2000 platform. The length of the complete mitochondrial genome was 15,685 bp, and the data were submitted to NCBI (MW770354). The genome contained 13 protein-coding genes (PCGs), 2 rRNA genes, 22 tRNA genes, and a putative control region. A phylogenetic tree was constructed based on the sequences of 13 PCGs identified by the maximum-likelihood method. Regardless of the lack of studies on the complete mitochondrial genome of other aquatic oligochaetes, the phylogenetic tree showed N. communis to cluster with Tubifex tubifex and Limnodrilus hoffmeisteri with high support value, and the freshwater oligochaete and earthworm groups to be sister groups.
The genus Nais Müller 1774 is a group of oligochaetes within the subfamily Naidinae, family Naididae Ehrenberg, 1828. These species are tiny worms that primarily inhabit freshwater, although some species are adapted to brackish environments (Brinkhurst and Jamieson Citation1971; Martínez-Ansemil and Prat Citation1984). Nais communis is a common species in this genus and appears to be more common than other Nais species in eutrophic waters (Dumnicka Citation1978; Bonacina et al. Citation1992; Juget and Lafont Citation1994; Jung Citation2011; Lee and Jung 2015). Therefore, as a representative species, N. communis is frequently used in ecological studies (Arimoro et al. Citation2007; Miserendino et al. Citation2008; Arslan and Mercan Citation2018). The morphological characteristics most frequently evaluated for differentiation of Nais species include the chaeta forms, i.e. ventral, hair chaetae, and needle chaetae. Differences between the chaetae of these species are often subtle and may overlap, leading to taxonomic confusion. This species also forms a species complex with many known cryptic species (Envall et al. Citation2012). For example, N. communis is morphologically similar to Nais variabilis Piguet, 1906, which often causes confusion in distinguishing them. The most frequently used morphological character to distinguish between them is the shape of their stomachs. The stomach of N. variabilis widens abruptly, whereas that of N. communis widens gradually (Brinkhurst and Jamieson Citation1971; Loden and Harman Citation1980; Envall et al. Citation2012). Since this is a minor difference, molecular studies of these two species are required to overcome the ambiguity. However, very few studies have reported the complete mitochondrial genomes of freshwater oligochaetes. In this study, we sequenced the mitogenome of N. communis and analyzed its phylogenetic position in the subclass Oligochaeta. We identified our specimen as N. communis because its anterior ventral chaetae were thinner and longer than the lower ones, with 4–5 chaetae per bundle. The needles had a clearly visible finely bifid with diverging teeth. Thus, our specimen had similar chaetae to morphotypes A3 and A4, which were regarded as N. communis lineages by Envall et al. (Citation2012). Further, it had a gradually widening stomach and could not swim when alive.
The specimen was collected on Jeju Island (Korea) in October 2019 (126° 51′ 21.42″E, 33° 49′ 55.14″N) and preserved in 80% ethanol; the voucher specimen was stored at the National Institute of Biological Resources (no. NIBRIV0000882545). Whole genomic DNA was extracted from posterior body segments of adult specimens using a REPLI-g Mitochondrial DNA Kit (Qiagen, Valencia, CA, USA). Whole-genome sequencing was performed using the Hiseq 2000 platform (Illumina). The mitochondrial genome was constructed using MITObim v1.9.1 (Hahn et al. Citation2013) and MITOS (Bernt et al. Citation2013). The sequence was deposited in GenBank (accession number MW770354). One new and 10 published mitochondrial genome sequences downloaded from GenBank, and Urechis caupo (Echiuroidea), included as an outgroup, were used for construction of the phylogenetic tree. Subsequently, annotations were performed using Geneious Prime 2019.2.1 (Kearse et al. Citation2012), and alignment was performed using MUSCLE Alignment. (Thompson et al. Citation2003). The best selected partitioning schemes and models of evolution were then obtained with ModelFinder (Kalyaanamoorthy et al. Citation2017), and a GTR + G + I model was identified as the best-fit model for the data. Maximum-likelihood analysis was conducted using PhyML 3.0 (Guindon et al. Citation2010) with 1000 bootstrap replicates.
The circular mitogenome of N. communis was 15,685 bp in size, with an overall base composition of 36.8% for A, 18.1% for C, 13.3% for G, and 31.8% for T. The genome exhibited codon biases with an AT content of 68.6% in protein-coding genes. The mitochondrial genome contained 13 protein-coding genes, two ribosomal RNA genes, and 22 tRNA genes. Of the 13 protein-coding genes, nine (ATP6, ATP8, COX1, COX2, COX3, CYTB, ND1, ND4, and ND6) used ATG as the start codon, two (ND2, ND3, and ND4L) used ATT as the start codon, and one (ND5) used ATA as the start codon.
Phylogenetic analysis, based on N. communis, of the mitogenomic sequences (13 PCGs) of all 12 species of annelids uploaded to GenBank indicated the relationships among groups within Annelida. Results showed that the newly sequenced species N. communis clustered together with Tubifex tubifex and Limnodrillus hoffmeisteri with high support value, indicating that freshwater oligochaete and earthworm groups are sister groups within Oligochaeta (). The relationships ((N. communis + T. tubifex + L. hoffmeisteri) + (Amynthas triastriatus + Metaphire californica + Metaphire guillelmi + Duplodicodrilus schmardae + Lumbricus rubellus + Drawida japonica)) + Chaetopterus variopedatus + Namalycastis abiuma + Urechis caupo were supported in Annelida.
Figure 1. Molecular phylogeny of N. communis (MW770354), one species in freshwater oligochaete, 10 species in annelids, and outgroup species based on complete mitogenome. The complete mitogenomes are downloaded from GenBank and the phylogenetic tree is constructed by the Maximum-likelihood method with 1000 bootstrap replicates.
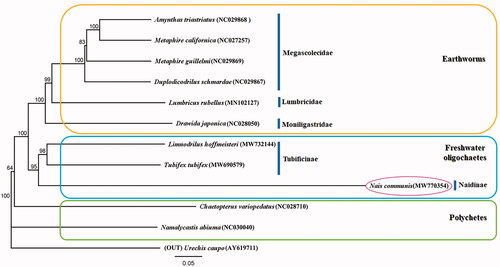
This study further clarified our understanding of the phylogenetic relationships of freshwater oligochaetes.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The genome sequence data that support the findings of this study are available in GenBank (https://www.ncbi.nlm.nih.gov) under accession no. MW770354. The associated data that support the findings of this study are also openly available in Mendeley Data at http://dx.doi.org/10.17632/7mfvhw5v87.1.
Additional information
Funding
References
- Arimoro FO, Ikomi RB, Iwegbue CM. 2007. Ecology and abundance of oligochaetes as indicators of organic pollution in an urban stream in southern Nigeria. Pak J Biol Sci. 10:446–453.
- Arslan N, Mercan D. 2018. Water quality determination of the lakes Mogan and Cernek (Turkey) by using macroinvertebrates accordance with water framework directive. Paper presented at: “UNITECH 2018” International Scientific Conference, 16-17 November 2018, Gabrovo, 510–518 pp.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Bonacina C, Bonomi G, Pasteris A. 1992. Some remarks on the macrobenthos community of the profundal zone of the large Italian lakes. Mem Inst Ital Idrobiol. 50:79–106.
- Brinkhurst RO, Jamieson BGM. 1971. Aquatic Oligochaeta of the world. Edinburgh: Oliver and Boyd. 860 pp.
- Dumnicka E. 1978. Communities of oligochaetes (Oligochaeta) of the River Nida and its tributaries. Acta Hydrobiol. 20:117–141.
- Envall I, Gustavsson LM, Erséus C. 2012. Genetic and chaetal variation in Nais worms (Annelida, Clitellata, Naididae). Zool J Linn Soc. 165(3):495–520.
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 59(3):307–321.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads—a baiting and iterative mapping approach. Nucleic Acids Res. 41(13):e129–e129.
- Juget J, Lafont M. 1994. Distribution of Oligochaeta in some lakes and pools of Bolivia. Hydrobiologia. 278(1-3):125–127.
- Jung JW. 2011. Naidid oligochaetes (Annelida: Clitellata) from the Seokhyeoncheon and Changreungcheon streams with new record of Nais variabilis. Korean J Limnol. 44:407–410.
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–589.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Lee JH, Jung JW, The Division of EcoCreative, Ewha Womans University. 2015. Six Korean new records of the Nais species (Annelida, Clitellata, Naididae). Korean J Environ Biol. 33(2):153–159.
- Loden MS, Harman WJ. 1980. Ecophenotypic variation in setae of Naididae (Oligochaeta). In: Brinkhurst RO, Cook DG, editors. Aquatic Oligochaeta biology. New York: Plenum Press; p. 33–39.
- Martínez-Ansemil E, Prat N. 1984. Oligochaeta from profundal zones of Spanish reservoirs. Hydrobiologia. 115(1):223–230.
- Miserendino ML, Brand C, Di Prinzio CY. 2008. Assessing urban impacts on water quality, benthic communities and fish in streams of the Andes Mountains, Patagonia (Argentina). Water Air Soil Pollut. 194(1-4):91–110.
- Thompson JD, Gibson TJ, Higgins DG. 2003. Multiple sequence alignment using ClustalW and ClustalX. Curr Protoc Bioinformatics Chapter 2(Unit 2):Unit 2.3.
