Abstract
The complete chloroplast genome of a staghorn fern species (Platycerium wallichii) was sequenced. The total genome was 158,286 bp in length, containing four regions: large single-copy (LSC) region (79,087 bp), small single-copy (SSC) region (21,397 bp), and two inverted repeat regions (IRs; 28,901 bp per each). In total 129 genes were annotated including 88 coding genes, 33 tRNAs, and 8 rRNAs. The overall GC content of the genome is 40.5%. Phylogenetic analysis supported the monophyly of both the subfamily Platycerioideae and the genus Platycerium. The genome data provides crucial information to support the future conservation and horticulture research.
Platycerium wallichii Hook. is an epiphytic fern belonging to the subfamily Platycerioideae of Polypodiaceae (PPGI 2016), it is distributed in tropical lowland rainforest, occurring in China, India, Malaysia, Myanmar, and Thailand (Zhang and Gilbert Citation2013). This species as well as its congeneric species is usually found on tree trunks forming a very impressively splendid view in the field, therefore Platycerium is popular ornamental fern in flower markets, which has resulted in an increasing loss of wild populations. Platycerium wallichii is considered to be endangered in China by deforestation and loss of habitat, it was classified as a national second-class protected wild plant in the Information System of Chinese Rare and Endangered Plants (ISCRPF) (http://www.iplant.cn/rep) and Critically Endangered by IUCN (https://www.iucn.org/resources/conservation-tools/iucn-red-list-threatened-species).
In this study, we report the complete chloroplast genome of P. wallichii for the first time. Fresh leaf material was collected from Yingjiang, Yunnan, China (24°45′01″N, 97°60′17″E). The specimen was deposited at the Herbarium of Xishuangbanna Tropical Botanical Garden, CAS (HITBC, http://www.xtbg.ac.cn/jgsz/zcxt/rdzwzzzyk/; Hongmei Liu, [email protected]) under the voucher number L2691/CP06). Genomic DNA was extracted from 2 g leaves using the CTAB method (Doyle and Doyle Citation1987), 0.5 ug DNA was fragmented to reconstruct short-insert (150 bp) libraries following the manufacturer’s manual (Illumina) and then used for sequencing. The DNA sample was indexed by tags and pooled together in one lane of a Genome Analyzer (Illumina HiSeq 2000) for sequencing at BGI-Shenzhen, and >4.0 Gb of reads were obtained. GetOrganelle toolkit (Jin et al. Citation2020) and Geneious (https://www.geneious.com) were employed to assemble and annotate the genome. The reference genomes identified as P. bifurcatum for annotation were MN623367 and MW042261. The newly sequenced and annotated plastid genome was submitted to the GenBank (accession number MW467509) and open to the public.
The chloroplast genome of P. wallichii has the quadripartite structure, with the total length is 158,286 bp including a large single-copy (LSC) region of 79,087 bp, a small single-copy (SSC) region of 21,397 bp, and a pair of inverted repeats regions (IR) of 28,901bp. GC content of the genome is 40.5%. The chloroplast genome contains 129 genes including 88 protein-coding genes, 33 tRNAs, and eight rRNA genes. Comparing the plastomes among the congeneric species, there is a sequence length variation within the genus with P. bifurcatum having both 160,442 bp (MW042261) and 156,985 bp (MN623367) in length with P. wallichii (158,286 bp) sitting in the middle.
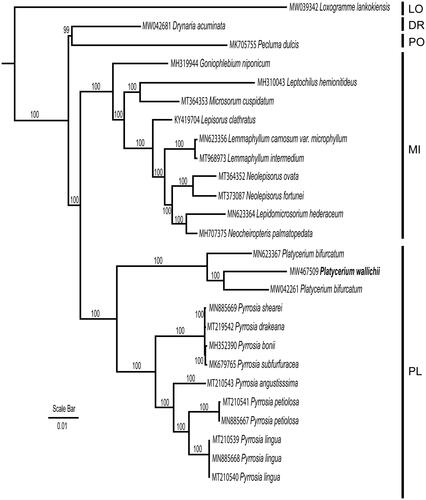
The genome sequence of P. wallichii was incorporated into a matrix including 26 taxa covering five subfamilies of Polypodiaceae (PPGI 2016) with the aim to reconstruct the infra-familial phylogenetic relationship. 80 coding genes were selected, aligned, and concatenated into a single matrix using MAFFT (Katoh and Standley Citation2013). The phylogenetic analyses were constructed using RAxML version 8.2.12 (Stamatakis Citation2014) with Maximum Likelihood method, nucleotide substitution model of GTR + Gamma was used with 1000 bootstrap replicates. Loxogramme lankokiensis (Loxogrammoideae) was selected as outgroup according to Schneider et al. (Citation2004). Monophyly of the subfamily Platycerioideae as well as the two genera Platycerium and Pyrrosia () was strongly supported. The genome of P. wallichii was nested between the two samples submitted as P. bifurcatum therefore we suspect one of the P. bifurcatum was misidentified.
Figure 1. Maximum likelihood phylogeny reconstructeded from 26 chloroplast genomes by RAxML. The sampling covered representatives of five subfamilies of Polypodiaceae. Loxogramme lankokiensis was selected as outgroup. LO: Loxogrammoideae; DR: Drynarioideae; PO: Polypodioideae; MI: Microsoroideae; PL: Platycerioideae.
The newly generated plastome of P. wallichii provides useful information for future phylogeny, conservation, and horticulture research, and will allow us to have a better understanding of the phylogenetic relationships for the derived fern family Polypodiaceae.
Disclosure statement
The authors are grateful to the open raw plastome data from public database. The authors are responsible for the content and have no potential conflict of interest.
Data availability statement
The genome sequence data of Platycerium wallichii that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/nuccore/MW467509.1) under the accession no. MW467509.. The associated Bioproject, Sequence Read Archive (SRA), and Biosample numbers are PRJNA737298, SRR14812826, and SAMN19689021, respectively.
Additional information
Funding
References
- Doyle JJ., Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Jin JJ, Yu WB, Yang JB, Song Y, de Pamphilis CW, Yi TS, Li DZ. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):241.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- PPG I. 2016. A community-derived classification for extant lycophytes and ferns. J Syst Evol. 54:563–603.
- Schneider H, Smith AR, Cranfill R, Hildebrand TJ, Haufler CH, Ranker TA. 2004. Unraveling the phylogeny of polygrammoid ferns (Polypodiaceae and Grammitidaceae): exploring aspects of the diversification of epiphytic plants. Mol Phylogenet Evol. 31(3):1041–1063.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Zhang XC, Gilbert MG. 2013. Platycerium. In: Wu ZY, Raven PH, Hong DY, eds. Flora of China. Vol. 2–3 (Pteridophytes). Beijing: Science Press; p. 796–797.
