Abstract
Using Oxford Nanopore and Illumina sequencing technologies, we reported the first complete mitochondrial genome of the important medicinal and edible plant Coriandrum sativum. The complete mitogenome was assembled into two circular-mapping forms of 82,926 bp (cir1) and 224,590 bp (cir2), respectively. There were 28 genes identified in the cir1 mitogenome, which included 14 protein-coding genes, 2 rRNA genes and 12 tRNA genes. There were 62 genes identified in the cir2 mitogenome, which included 41 protein-coding genes, 5 rRNA genes and 16 tRNA genes. Phylogenetic analysis showed that Coriandrum sativum was most closely related to Daucus carota.
Coriandrum sativum (Coriander), a member of the Apiaceae family, belongs to the Mediterranean aromatic herb and is widely cultivated in the world (Tulsani et al. Citation2020). The coriander leaves, stems and roots with a large number of bioactive compounds of coriander are all edible, which are usually used as vegetable, spice and medicine (Sahib et al. Citation2013). Samples of Coriandrum sativum were obtained from Tianjin Academy of Agricultural Sciences (18°13′47.45″N, 109°30′55.29″E, TAAS) in China. The specimen was deposited at Biotechnology Research Institute of TAAS (voucher number: HB202015). In this study, we used the Oxford Nanopore Technologies (ONT) combined with Illumina Hiseq system to sequence the first complete mitochondrial genome of C. sativum (Kan et al. Citation2020).
The complete mitogenome of Coriandrum sativum was assembled into two circular-mapping forms of 82,926 bp (cir1) and 224,590 bp (cir2) in length by the SPAdes assembler (Bankevich et al. Citation2012), which deposited under the GenBank accession numbers (MW477237 and MW477238). In comparison with other mitogenomes in Apiaceae, the mitogenome of C. sativum (∼83kb/∼224kb) was smaller than that of D. carota (∼281kb) and B. falcatum (∼464kb) (Iorizzo et al. Citation2012; Kim et al. Citation2020). The mitochondrial base composition of cir1 and cir2 were A 27.25%, T 27.92%, C 22.66%, G 22.17% and A 27.44%, T 27.41%, C 22.88%, G 22.27%, respectively. There were 28 genes identified in the cir1 mitogenome, which included 14 protein-coding genes, 2 rRNA genes and 12 tRNA genes. There were 62 genes identified in the cir2 mitogenome, which included 41 protein-coding genes, 5 rRNA genes and 16 tRNA genes. Only ten genes contained intron, including eight protein-coding genes (nad1, nad2, nad4, nad5, nad7, rps3, ccmFc, and cox2), and two tRNA (trnA-UGC and trnP-CGG). Comparison between the C. sativum and D. carota mitogenomes, showed that only one unique gene (ycf2) was not observed in the latter. The function of ycf2 gene is unknown, and the similar result appeared in the Mirabilis himalaica mitogenome (Yuan et al. Citation2020).
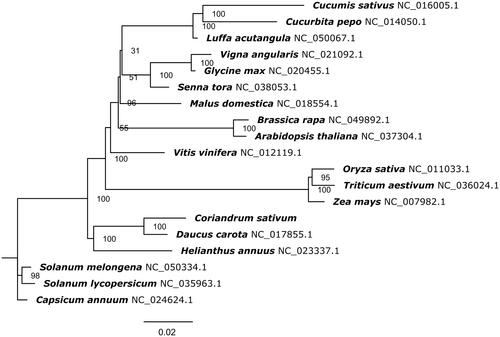
The phylogenetic tree was constructed by using RAxML (v8.2.9) with 1,000 bootstrap replicates (Stamatakis Citation2014). A maximum likelihood analysis was performed on nineteen species based on the conserved sequences of the whole mitochondrial genome. The phylogenetic tree showed that C. sativum was most related to D. carota with 100 bootstrap values (). These results will contribute to a better understanding of Apiaceae evolution and further development of molecular markers in Coriandrum sativum used for germplasm classification.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at [https://www.ncbi.nlm.nih.gov] (https://www.ncbi.nlm.nih.gov/) under the accession no.MW477237-MW477238. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA729412, SUB3752698, and SAMN19115529 respectively.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Iorizzo M, Senalik D, Szklarczyk M, Grzebelus D, Spooner D, Simon P. 2012. De novo assembly of the carrot mitochondrial genome using next generation sequencing of whole genomic DNA provides first evidence of DNA transfer into an angiosperm plastid genome. BMC Plant Biol. 12:61.
- Kan SL, Shen TT, Gong P, Ran JH, Wang XQ. 2020. The complete mitochondrial genome of Taxus cuspidata (Taxaceae): eight protein-coding genes have transferred to the nuclear genome. BMC Evol Biol. 20(1):10.
- Kim CK, Jin MW, Kim YK. 2020. The complete mitochondrial genome sequences of Bupleurum falcatum (Apiales: Apiaceae)). Mitochondrial DNA B Resour. 5(3):2576–2577.
- Sahib NG, Anwar F, Gilani AH, Hamid AA, Saari N, Alkharfy KM. 2013. Coriander (Coriandrum sativum L.): a potential source of high-value components for functional foods and nutraceuticals-a review. Phytother Res. 27(10):1439–1456.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Tulsani NJ, Hamid R, Jacob F, Jacob F, Umretiya NG, Nandha AK, Tomar RS, Golakiya BA. 2020. Transcriptome landscaping for gene mining and SSR marker development in coriander (Coriandrum sativum L.). Genomics. 112(2):1545–1553.
- Yuan F, Xu YJ, Lan XZ. 2020. The complete mitochondrial genome sequence of Mirabilis himalaica, an endemic plant to Tibet. Mitochondrial DNA B Resour. 5(3):2802–2804.