Abstract
Potentilla lineata Treviranus is a common medicinal plant distributed in the southwest of China. In this study, we sequenced the complete chloroplast genome sequence of P. lineata and investigated its phylogenetic relationship within Potentilla and related genera. The total length of the chloroplast genome is 156,985 bp. The genome exhibits a typical quadripartite structure containing a pair of IRs (inverted repeats) of 25,974 bp separated by a small single copy (SSC) region of 18,859 bp and a large single copy (LSC) region of 86,178 bp. The chloroplast genome contained 112 genes, including 78 protein-coding genes, 30 tRNA genes, and four rRNA genes. The phylogenetic analysis indicates that Potentilla L. is a monophytic taxon that is sister to Potaninia Maxim. Potentilla lineata is closely related to P. centigrana Maxim in the present study. This study provides a reference for the phylogeny and species evolution of the genus Potentilla and related genera.
Potentilla lineata Treviranus is classified in a complex genus with about 500 species, namely Potentilla in the Rosaceae (Xu and Podlech Citation2010). The species is mainly distributed in Yunnan, Guizhou and Tibet provinces of China. As a common medicinal plant, the root of P. lineata is used to treat enteritis and diarrhea, traumatic bleeding, and is an anti-inflammatory (Manju et al. Citation2006). The plant is widely used by Bai and Dai people due to its medicinal value (Duan Citation2017; Jiang Citation2017). Studies on this species have mainly focused on pharmacological effects and active ingredients (Chen et al. Citation2013; Toğrul et al. Citation2015). However, the chloroplast genome of P. lineata has not been analyzed. Here we sequenced the complete chloroplast genome sequence of P. lineata, as well as reconstructed a phylogenetic tree with other taxa classified in the Rosaceae to determine its evolutionary history.
Fresh and cleaned leaf materials of P. lineata were sampled from Dali, Yunnan, China (N 25°52′38.91″, E 100°00′27.25″). A voucher specimen (No. ZDQ021) was collected and deposited at the Herbarium of Medicinal Plants and Crude Drugs of the College of Pharmacy, Dali University (Professor Dequan Zhang, [email protected]). The total genomic DNA was extracted using an improved CTAB method (Yang et al. Citation2014), and sequenced with Illumina Hiseq 2500 (Novogene, Tianjing, China) platform using a pair-end (2 × 300 bp) library. The raw data were filtered using Trimmomatic v.0.32 with the default settings (Bolger et al. Citation2014). The resulting paired-end reads were assembled into a circular contigs using GetOrganelle.py (Jin et al. Citation2020).The complete chloroplast genome was annotated with Geneious 8.0.2 software (Kearse et al. Citation2012) and the annotated results were obtained after manual correction.
The annotated chloroplast genome was submitted to the GenBank with an accession number MT677853. The complete chloroplast genome was 156,985 bp. It consists of a typical quadripartite structure with a pair of IRs (inverted repeats) of 25,974 bp that are separated by a small single copy (SSC) region of 18,859 bp and a large single copy (LSC) region of 86,178 bp. The overall GC content is 36.7%, while 34.4%, 30.5%, and 42.7% are respectively calculated for the LSC, SSC, and IR regions. The chloroplast genome contains 112 genes, including 78 protein coding, 30 tRNA, and four rRNA genes. Of these, 17 genes occur in duplicate in the inverted repeat regions, 9 genes (rpl2, rpl16, rps12, rps16, rpoC1, ndhB, ndhA, petD, petB) contained one intron, while two genes (ycf3 and clpP) have two introns.
Figure 1. Phylogenetic tree constructed using BI method based on chloroplast genome sequences of 22 species in Rosoideae. (Note: the number on each node represents the posterior probability).
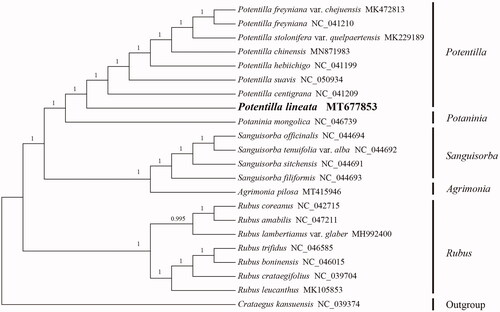
For the phylogenetic analysis, 21 complete chloroplast genome sequences of Rosoideae were downloaded from the NCBI database. The sequences were aligned with MAFFT V.7.149 using the default settings (Katoh and Standley Citation2013). The nucleotide substitution model was determined by jModelTest v.2.1.7 (Darriba et al. Citation2012). A Bayesian inference (BI) analysis was performed by MrBayes v.3.2.6 (Ronquist et al. Citation2012) with Crataegus kansuensis Wils. (No. NC_039374) designated as the outgroup (). The phylogenetic analysis indicated Potentilla L. is a monophyletic genus, which is consistent with the previous phylogenetic analysis by Faghir et al. (Citation2014) Li et al. (Citation2020) and Li et al. (Citation2021). In this analysis, Potentilla is sister to Potaninia Maxim. Potentilla lineata was fully resolved in a clade with P. centigrana Maxim., P. suavis and P. hebiichigo . This complete chloroplast genome of P. lineata provides a reference for further phylogenetic studies and the evolution of species in Potentilla and related genera, as well as for conservation and utilization of the resources.
Disclosure statement
The authors declare no conflicts of interest and are responsible for the content.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. MT677853. The associated BioProject, SRA, and BioSample numbers are PRJNA737049, SRR14825541, and SAMN19698019 respectively.
Additional information
Funding
References
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30(15):2114–2120.
- Chen X, Jiang ZT, Li R. 2013. Purification and HPLC-ESI-MS/MS analysis of Flavonoids from Potentilla lineata. Mod Food Sci Technol. 29(12):3031–3037.
- Darriba D, Taboada GL, Doallo R, Posada D. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 9(8):772.
- Duan BZ. 2017. Journal of Dai medicine. Kunming: Yunnan Science and Technology Press; p. 131
- Faghir MB, Attar F, Farazmand A, Osaloo SK. 2014. Phylogeny of the genus potentilla (Rosaceae) in Iran based on nrDNA ITS and cpDNA trnL-F sequences with a focus on leaf and style characters' evolution. Turk J Bot. 38:417–429.
- Jiang B. 2017. Atlas of medicinal plants of Bai nationality. Beijing: China traditional Chinese Medicine Press; p. 15.
- Jin JJ, Yu WB, Yang JB, Song Y, dePamphilis CW, Yi TS, Li DZ. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):241.
- Katoh K, Standley D. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kearse M, Moir R, Wilson A, Stones HS, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Li Y, Li X, Ding FB, Li Q, Yan HL, Li RN, Zhang L, Wang H, Zhao Y. 2021. The complete chloroplast genome sequence of Potentilla tanacetifolia Willd. ex Schltdl. Mitochondrial DNA B Resour. 6(2):478–479.
- Li QQ, Zhang ZP, Ha SBG. 2020. The complete chloroplast genome of Potentilla suavis (Rosaceae: Potentilleae). J Inner Mongolia Normal Univ Nat Sci ed. 49(6):471–474.
- Manju S, Anjali D, Sushma K, Ashok A. 2006. Rapid in vitro propagation of Potentilla fulgens wall—a Himalayan alpine herb of medicinal value. J Plant Biochem Biotechnol. 15(2):143–145.
- Ronquist F, Teslenko M, Mark P, Ayres DL, Darling A, H€Ohna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542.
- Toğrul C, Balsak D, Ekinci C, Seçkin KD, Ekinci A, Tahaoğlu AE, Bademkiran H, Görkem U, Görük NY, Güngör T, et al. 2015. Effects of Potentilla fulgens as a prophylactic agent for ischemia/reperfusion injury in the rat ovary. Anal Quant Cytopathol Histpathol. 37(5):310–316.
- Xu LR, Podlech D. 2010. Potentilla L. In: Wu ZY, Raven PH, editors. Flora of China. Beijing: Science Press; St. Louis: Missouri Botanical Garden Press; p. 233–244.
- Yang JB, Li DZ, Li HT. 2014. Highly effective sequencing whole chloroplast genomes of angiosperms by nine novel universal primer pairs. Mol Ecol Resour. 14(5):1024–1031.
