Abstract
Illigera grandiflora, a kind of traditional medicinal liana, belongs to the Illigera Blume of the Hernandiaceae. In this study, we reported the characteristics of complete plastome for I. grandiflora. Its total plastome was 156,138 bp in length, comprising a large single-copy region(LSC) of 84,931 bp, a small single-copy region (SSC) of 18,544 bp, and a pair of inverted repeat (IR) regions of 26,549 bp. The overall GC content was 39.16% (LSC, 37.77%; SSC, 33.89%; IR, 43.21%). The plastome encoded 134 genes, including 83 protein-coding genes, 42 transfer RNA genes, and 10 ribosomal RNA genes. The relationships in our phylogeny showed that the two Illigera species are located in the same clade, with Hernandia nymphaeifolia being the next sister group, followed by Wilkiea huegeliana.
Illigera grandiflora W.W.Sm. & Jeffrey, an evergreen liana with 2–6 m tall, inhabits forests at an altitude of 800–2100 m and is widely distributed in India, north Myanmar, and southwestern China (Chinese Flora Editorial Board, Chinese Academy of Sciences Citation2008). The root and stem of I. grandiflora was used to treat dropsy and traumatic injury (Gao Citation2007; Huang Citation1985), and previous studies have revealed the major chemical components of the plants of the genus Illigera Blume are alkaloids and terpenoids. Li et al. (Citation2019) have isolated a new dibenzopyrrocoline alkaloid, together with five known ones from I. grandiflora, three of them exhibited the moderate inhibitory activity against acetylcholinesterase (AChE) or butyrylcholinesterase (BChe), it has showed great medical potential. But there are few studies of I. grandiflora on genomic at present. Recent studies (Xin et al. Citation2020) have finished the complete chloroplast genomes sequencing of Illigera celebica (LAU199). In order to compare the chloroplast differences among different species and to better understand its phylogenetic relationships between them and other Laurales species, we reported the characteristics of complete plastome for I. grandiflora, and then reconstructed a phylogenetic tree.
Fresh leaves of I. grandiflora were collected from Cangyuan Wa Autonomous County, Yunnan, China: (23.2°N, 99.4°E) for genomic DNA extraction using modified CTBA method (Cai et al. Citation2014), and was then sequenced by Illumina Hiseq 2000 platform at BGI-Shenzhen. A specimen was deposited at XTBG’s Biodiversity Research Group (contact Song-Yu, [email protected]) under the voucher number SY5852. Aligning, assembly (reference sequences are LAU00199 and MN990581), and annotation were conducted by MAFFT v.7 online program (https://mafft.cbrc.jp/alignment/server/) (Katoh and Standley Citation2013), GetOrganelle software (Jin et al. Citation2018), and Geneious R8.1.3 (Kearse et al. Citation2012) respectively. The plastid genome phylogenetic relationships were reconstructed based on a maximum-likelihood (ML) analysis with the GTR + F + R2 model by iqtree version 1.6.7.1 program using 1000 bootstrap replicates (Nguyen et al. Citation2015).
The plastome of I. grandiflora (MW755975) is a circular DNA molecule with a length of 156,138 bp, which is 15 bp larger than Illigera celebiea (LAU00199). The complete plastome contains a large singlecopy region (LSC, 84,931 bp), a small single-copy region (SSC, 18,544 bp), and a pair of inverted repeats (IRs, 26,549 bp). The overall GC content is 39.16%, the corresponding values of the LSC, SSC,and IR regions are 37.77%, 33.89%, and 43.21% respectively. The plastome encoded a set of 120 genes, of which 76 are protein‐coding genes, 36 are transfer RNA genes, and 8 are rRNA genes.
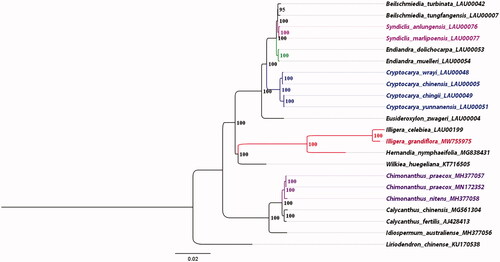
The ML tree was built on complete plasomes of 22 related species, Liriodendron chinense (KU170538) was treated as the out-group (). The phylogenetic tree was divided into three mian clades corresponding to four families: Hernandiaceae, Monimiaceae, Lauraceae, and Calycanthaceae. Most relationships had high internal support. Phylogenetic analysis based on all plastomes supported that sisterhood of I. grandiflora and I. celebiea, with H. nymphaeifolia (MG838431) being the next sister group, followed by W. huegeliana (KT716505). In addition, we reconfirmed the sisterhood of Lauraceae and a clade containing Hernandiaceae and Monimiaceae (Song et al. Citation2020).
Figure 1. The ML phylogenetic tree for I. grandiflora based on other 21 species (11 in Lauraceae, 2 in Hernandiaceae, 1 in Monimiaceae, 6 in Calycanthaceae, and 1 in Magnoliaceae) plastid genomes; the complete plastome sequences were from Lauraceae Chloroplast Genome Database (https://lcgdb.wordpress.com/) (13 species those numbers ending with LAU) and NCBI (other 9 species).
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. MW755975. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA715651, SAMN18388833, and SRR14018806, respectively.
Additional information
Funding
References
- Cai WJ, Xu DB, Lan X, Xie HH, Wei JG. 2014. A new method for the extraction of fungal genomic DNA. Agric Res Appl. 152:1–5.
- Chinese Flora Editorial Board, Chinese Academy of Sciences. 2008. Flora of China. Vol. 7. J B Sci Press(Beijing); p. 1153–1926.
- Gao Z. 2007. It is made up of plant extracts such as Sinomenium spp., Aconitum carmichaeli Debx. and Paeonia lactiflora Pall to treat arthritis and pain. Drugs Clin. 22 (04):181–182.
- Huang X. 1985. A new species and medicinal plant of the same genus from Illigera BI of Guangxi. Guihaia. 5(1):17–20.
- Jin JJ, Yu WB, Yang JB, Song Y, Yi TS, Li DZ. dePamphilis CW, Yi TS, Li DZ. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biology. 21(1):241.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Li XJ, Dong JW, Gan D, Zhou DJ, Cai XY, Cai L, Ding ZT. 2019. (-)-Grandiflorimine, a new dibenzopyrrocoline alkaloid with cholinesterase inhibitory activity from Illigera grandiflora. Nat Prod Res. 35(5):763–769.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Song Y, Yu WB, Tan YH, Jin JJ, Wang B, Yang JB, Liu B, Corlett RT. 2020. Plastid phylogenmics improve phylogenetic resolution in the Lauraceae. J Syst Evol. 58(4):423–439.
- Xin YX, Xin J, Yao GQ, Qu YY, Feng FY, Song Y, Sun ZH. 2020. The complete chloroplast genome sequence of Illigera celebica. Mitochondrial DNA B Resour. 5(3):2454–2455.
