Abstract
Choerospondias axillaris, an ancient and versatile plant of Anacardiaceae, which is widespread in eastern Asia. The complete chloroplast (cp) genome of C. axillaris was generated and determined in this work. The genome of cp was 159,131 bp in length with a total GC content of 37.6%. The circular molecular genome presented a quadripartite structure, comprising a 91,305 bp large single-copy region (LSC), a 19,092 bp small single-copy region (SSC), and a pair of inverted repeat regions with 24,367 bp. The complete genome encoded 128 genes, including 83 protein coding, 37 tRNA and eight rRNA genes. Phylogenetic analysis indicated that C. axillaris was most related to Sclerocarya birrea.
Choerospondias axillaris (Roxb.) B. L. Burtt et A. W. Hill is a deciduous broad-leaved tree species of Anacardiaceae, which is a monospecific species of genus Choerospondias in eastern Asia and mainly distributed in Japan and South China. C. axillaris has a long cultivation and utilization history, whose fruit fossil with 15-million-year-old was first discovered in Fujian, China (Wang et al. Citation2020). For its characteristics of fast growth and strong adaptability, C. axillaris is suitable for artificial afforestation. So that C. axillaris has a growing popularity and important economic effects, it is noted for timber, edible, and medicinal values. Previous researches on C. axillaris have mainly focused on pharmacological action (Sun et al. Citation2015; Mann et al. Citation2020), phytochemical investigation (Li et al. Citation2018), photosynthetic physiology (Li and Xu Citation2020), and morphological diversity (Wang et al. Citation2019). However, there is no report on the chloroplast (cp) genome of C. axillaris. The complete cp genome has been progressively analyzed in diverse species to provide insight into species identification and population dynamics (Shaw et al. Citation2014). In this study, we established and characterized the complete C. axillaris cp genome and performed phylogenetic analysis with the cp genomes of other species of Anacardiaceae.
The leaves of C. axillaris were sampled from Chenshan Botanical Garden, Shanghai, China (31°08′N, 121°18′E). And the voucher specimen (Accession number: CSSH202104) was stored in Shanxi Datong University (http://www.sxdtdx.edu.cn/, Kun Zhang, [email protected]). Genomic DNA extraction of C. axillaris was processed according to the modified CTAB method (Murray and Thompson Citation1980). After DNA purification, the libraries with an average length of 350 bp were constructed using the NexteraXT DNA Library Preparation Kit (Illumina, San Diego, CA), and high-throughput sequencing was carried out on Illumina Novaseq 6000 platform. In total, 3.7 Gb clean reads were generated by editing raw sequence reads with NGS QC Tool kit (Patel and Jain Citation2012). The cp genome was de novo assembled by SPAdes version 3.11.0 software (Bankevich et al. Citation2012), and cp genes of C. axillaris were annotated by using PGA (Qu et al. Citation2019) through aligning to genome of Sclerocarya birrea (A. Rich.) Hochst cp (Accession number: NC043919). The annotated sequence was submitted to GenBank and data were openly available at (https://www.ncbi.nlm.nih.gov/nuccore/MZ042936.1/) under the accession MZ042936. The associated SRA number is SRR14328382.
The cp genome of C. axillaris was a circular molecular genome presented a quadripartite structure. The genome was 159,131 bp in length, and consisted of a 91,305 bp large single-copy (LSC) region, a 19,092 bp small single-copy (SSC) region, and a pair of inverted repeat (IRa and IRb) regions with 24,367 bp. The overall GC content detected in the C. axillaris cp genome was 37.6%. The assembled genome encoded 128 genes, including 83 protein coding, 37 tRNA and eight rRNA genes. The majority of these genes did not contain intron, while 15 genes (atpF, ndhA, ndhB, petB, petD, rpl2, rpl16, rpoC1, rps16, trnA-UGC, trnG-UCC, trnI-GAU, trnK-UUU, trnL-UAA, and trnV-UAC) contained one intron and two genes (clpP and ycf3) contained double introns. All genes occurred as a single copy, except that 10 genes (ycf1, rrn4.5, rrn5, rrn16, rrn23, tRNA-Ile, tRNA-Ala, tRNA-Arg, tRNA-Asn, and ORF302) were duplicated in IR regions.
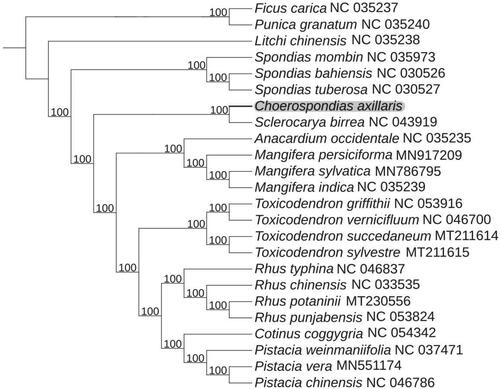
To further determine the phylogenetic position of C. axillaris, 20 complete cp genomes of different species within Anacardiaceae family and other three species as outgroup were selected to construct a phylogenetic tree. The sequences were downloaded from NCBI GenBank database and aligned by using MAFFT (Katoh and Standley Citation2013). Subsequently, a maximum-likelihood phylogenetic tree was established by IQTREE version 1.6 (Jana et al. Citation2016) with 1000 bootstrap replicates. The main genera displayed on the phylogenetic tree were Rhus, Mangifera, Toxicodendron, Pistacia, and Spondias. Phylogenetic analysis revealed that C. axillaris exhibited the closest relationship with Sclerocarya birrea (). The information derived from this work provides a basis for future genetic and evolutionary studies in C. axillaris, which may help facilitate the utilization and protection of this ancient and versatile plant.
Figure 1. Maximum-likelihood phylogenetic tree for C. axillaris based on 21 complete cp genome in Anacardiaceae, with Litchi chinensis Sonn., Punica granatum L., and Ficus carica L. as outgroup. The bootstrap values are located on each node and the Genbank accession numbers are shown beside the Latin name of the species.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the article.
Data availability statement
The assembled complete cp genome sequence of C. axillaris has been submitted to GenBank of NCBI and is openly available under the accession number: MZ042936 (https://www.ncbi.nlm.nih.gov/nuccore/MZ042936.1/). The associated BioProject, SRA, and Bio-Sample numbers are PRJNA725317, SRR14328382, and SAMN18875962, respectively.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Jana T, Lam-Tung N, Arndt VH, Quang MB. 2016. W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 44:W232–W235.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Li GY, Xu JM. 2020. Effects of light and season on chlorophyll fluorescence parameters of Choerospondias axillaris families. Mol Plant Breeding. 18(12):4097–4104.
- Li Q, Liu CM, Li T, McClements DJ, Fu YX, Liu JY. 2018. Comparison of phytochemical profiles and antiproliferative activities of different proanthocyanidins fractions from Choerospondias axillaris fruit peels. Food Res Int. 113:298–308.
- Mann S, Sharma A, Sarkar A, Kharb R, Malhotra R, Datta B, Gupta RK, Biswas S. 2020. Evaluation of anti-inflammatory effects of Choerospondias axillaris fruit’s methanolic extract in Synoviocytes and CIA rat model. Curr Pharm Biotechnol. 21(7):596–604.
- Murray MG, Thompson WF. 1980. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 8(19):4321–4326.
- Patel RK, Jain M. 2012. NGS QC toolkit: a toolkit for quality control of next generation sequencing data. PLoS One. 7(2):e30619.
- Qu XJ, Moore MJ, Li DZ, Yi TS. 2019. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods. 15(1):50–61.
- Shaw J, Shafer HL, Leonard OR, Kovach MJ, Schorr M, Morris AB. 2014. Chloroplast DNA sequence utility for the lowest phylogenetic and phylogeographic inferences in angiosperms: the tortoise and the hare IV. Am J Bot. 101(11):1987–2004.
- Sun B, Xia QM, Gao ZY. 2015. Total flavones of Choerospondias axillaris attenuate cardiac dysfunction and myocardial interstitial fibrosis by modulating NF-κB signaling pathway. Cardiovasc Toxicol. 15(3):283–287.
- Wang XA, Wei XX, Wu RJ, Ye XF. 2019. Morphological diversity of 49 Choerospondias axillaris germplasms in Fujian. Fujian J Agri Sci. 34(4):400–408.
- Wang ZX, Herrera F, Shu JW, Yin SX, Shi GL. 2020. A new Choerospondias (Anacardiaceae) endocarp from the middle Miocene of Southeast China and its paleoecological implications. Rev Palaeobot Paly. 283:104312.
