Abstract
Agriocnemis femina (Brauer, 1868) and Ischnura senegalensis (Rambur, 1842) are two damselflies inhabiting paddy lands. As an intermediate predator, they play an important role in controlling certain crop pest and mosquitoes. In this study, we sequenced complete mitogenomes of these two species. The total length of mitogenomes is 15,936 bp in A. femina and 15,762 bp in I. senegalensis. Both of mitogenomes consist of 13 protein-coding genes, 22 tRNA genes, two rRNA genes, and one control region. The close relationship between I. senegalensis and I. elegans was further proved by phylogenetic analysis. Our phylogenetic analysis indicated a clear two lineages in Coenagrionidae (Core and ridge-faced Coenagrionidae). Ridge-faced Coenagrionidae consisted of Megaloprepus caerulatus and Ceriagrion fallax. In core Coenagrionidae, Ischnura and Enallagma are most closely related; they formed one clade with Agriocnemis and then grouped together with Paracerion. Our study provides new genetic information for further study in phylogenetic analysis of Coenagrionidae.
Odonates represent a prime group of non-model organisms that are widely used to study and answer key questions in ecology and evolution (Zaka et al. Citation2014). For instance, odonates are amongst the most ancient groups of insects and own a crucial position in the evolution of winged insects (Bechly Citation1995). In suborder Zygoptera, Coenagrionidae includes more than 1100 species, which is a diverse damselfly family with worldwide distribution. In this study, we sequenced and characterized two mitogenomes from the damselfly family Coenagrionidae (Agriocnemis femina, Brauer, 1868 and Ischnura senegalensis, Rambur, 1842). Both of these two species inhabit paddy land and become important predators for biological control of crop pests or mosquitoes (Kandibane et al. Citation2005).
The adult damselflies of A. femina (Brauer, 1868) and I. senegalensis (Rambur, 1842) were collected by sweep net in the Shenshan park of Wuhu, China (118°24’53” E, 31°20’51” N for A. femina and 118°24’57” E, 31°20’58” N for I. senegalensis) in July, 2019 and preserved in 90%-ethanol at room temperature. The specimen was deposited at Entomological Evolution and Ecology Room in Anhui Normal University (Bin Jiang, [email protected]) under the voucher number L502-SS03. Genomics DNA was extracted from muscle tissues of thorax using CTAB methods (Bechly Citation1995). The quality of DNA was checked by using NanoDrop and measured by Qubit. Samples were sent for library preparation and paired-end sequencing with Novaseq following the standard protocol from Illumina in BenaGen Inc. (Wuhan, China). The adapters and low-quality reads in raw data were trimmed and filtered by using SOAPnuke 1.3.0. Genome assemblies were conducted using SPAdes 3.13.0 (parameter: -k 127) (Dierckxsens et al. Citation2016). We used MITOS web server (http://mitos.bioinf.uni-leipzig.de) to annotate genomes (Bernt et al. Citation2013).
The total length of the new mitogenomes in our study is 15,936 bp in A. femina (Accession number in Genbank: MT787566) and 15,762 bp in I. senegalensis (Accession number in Genbank: MT787567). We noticed that a partial mitogenome of A. femina (13,280 bp, accession number in Genbank: MK951667) is reported earlier. Comparing MK951677, 20 SNPs were found; one T-deletion in the interspace between trnT and trnP and one A-deletion and two A-insertions in 16S RNA coding region in MT787566 were found. Like other mitogenomes in Coenagrionidae, both new mitogenomes from us contain 13 protein-coding genes, 22 tRNA genes, two rRNA genes, and one control region. All tRNA sequences length range from 63 to 72 bp. In both mitogenomes, the shortest tRNA sequence is trnG and the longest one is trnK. According to the predicted tRNA structure from MITOs, all tRNA sequences can form the characteristic clover leaf secondary structures. In both mitogenomes, trnS1 lacks DHU-stem and trnF lacks TΨC loop. For protein-coding genes, in A. femina eight genes (cox1, cox2, atp6, cox3, nad3, nad4, nad4L, and cob) use ATG/ATA (encoding for methionine) and 5 genes (nad2, atp8, nad5, nad6 and nad1) use ATT (encoding for isoleucine) as start codon; in I. senegalensis, nad6 use ATA and nad3 use ATT as start codon and the other genes are the same as in A. femina. We identified a standard stop codon for most of protein-coding genes; however, cox3 and nad5 in A. femina and nad5 in I. senegalensis had an incomplete stop codon.
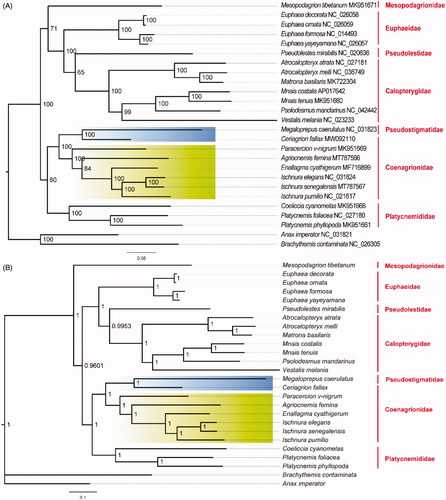
In order to determine the phylogenetic relationship of A. femina and I. senegalensis in Coenagrionidae, we collected 22 other mitogenome sequences from GenBank in Zygoptera (Lin et al. Citation2010; Lorenzo-Carballa et al. Citation2014; Chen et al. Citation2015; Feindt et al. Citation2016; Okuyama and Takahashi Citation2017; Zhang et al. Citation2017; Xu et al. Citation2018; Lan et al. Citation2019; Song et al. Citation2019; Wang et al. Citation2019; Shao et al. Citation2021) and two outgroup species from Anisoptera (Anax imperator (Herzog et al. Citation2016); Brachythemis contaminata (Yu et al. Citation2016)). We reconstructed the Maximum-likelihood tree by iqtree 2.1.2 (Nguyen et al. Citation2015) (). To assess ML nodal support, we calculated 1000 likelihood bootstrap replications. Bayesian analyses were conducted with MrBayes v3.2 (Ronquist et al. Citation2012). The GTR + I+G substitution model was identified as the best model by Modeltest 3.7 (Posada & Crandall Citation1998). MCMC were performed for 100,000 generations and terminated after the average split frequencies falling below 0.01. Trees were sampled every 100 replicates with the first 25% of samples discarded as burn-in. Both phylogeny analyses indicated that Coenagrionidae separated into core Coenagrionidae and ridge-faced Coenagrionidae two lineages (Dijkstra et al. Citation2014). Newly sequenced I. senegalensis has a close phylogenetic relationship with I. elegans and forms one clade with I. pumilio. The close relationship between I. elegans and I. senegalensis can also be proved by their interspecific hybridization in the lab (Okude et al. Citation2020). Ischnura species formed one clade with Enallagma species and then clustered with newly sequenced A. femina. Therefore, the complete mitogenomes of I. senegalensis and A. femina in Coenagrionidae provide valuable genetic information for further phylogenetic analyses in Zygoptera.
Figure 1. Phylogenetic relationships based on 13 mitochondrial protein-coding genes in Zygoptera. Branches with green background indicate core Coenagrionidae and branches with blues background represent ridge-faced Coenagrionidae. Nodal values indicate (A) bootstrap support values in ML tree and (B) the posterior probabilities in Bayes tree.
Data available statement
The data that support the findings of this study are openly available in NCBI (National Center for Biotechnology Information) at https://www.ncbi.nlm.nih.gov/, reference number MT787566 and MT787567. The associated BioProject, SRA, and Bio-Sample numbers of Ischnura senegalensis are PRJNA730433, SRR14574107, and SAMN19231599. For Agriocnemis femina, submission numbers are successively PRJNA730438, SRR14574586 and SAMN19231647.
Disclosure statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Additional information
Funding
References
- Bechly G. 1995. Morphological analysis of the wing venation of extant dragonflies and their stem group representatives (Insecta; Pterygota; Odonata) with special reference to phylogenetic systematics and the ground plan of crown group Odonata. Petalura. 1:341.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Chen M-Y, Chaw S-M, Wang J-F, Villanueva RJT, Nuñeza OM, Lin C-P. 2015. Mitochondrial genome of a flashwing demoiselle, Vestalis melania from the Philippine Archipelago. Mitochondrial DNA. 26(5):720–721.
- Dierckxsens N, Mardulyn P, Smits G. 2016. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Dijkstra K-DB, Kalkman VJ, Dow RA, Stokvis FR, Van Tol JAN. 2014. Redefining the damselfly families: a comprehensive molecular phylogeny of Zygoptera (Odonata). Syst Entomol. 39(1):68–96.
- Feindt W, Osigus H-J, Herzog R, Mason CE, Hadrys H. 2016. The complete mitochondrial genome of the neotropical helicopter damselfly Megaloprepus caerulatus (Odonata: Zygoptera) assembled from next generation sequencing data. Mitochondrial DNA B Resour. 1(1):497–499.
- Herzog R, Osigus HJ, Feindt W, Schierwater B, Hadrys H. 2016. The complete mitochondrial genome of the emperor dragonfly Anax imperator LEACH, 1815 (Odonata: Aeshnidae) via NGS sequencing. Mitochondrial DNA B Resour. 1(1):783–786.
- Kandibane M, Raguraman S, Ganapathy N. 2005. Relative abundance and diversity of Odonata in an irrigated rice field of Madurai, Tamil Nadu. Zoos Print J. 20(11):2051–2052.
- Lan D-Y, Shen S-Q, Cai Y-Y, Wang J, Zhang J-Y, Storey KB, Yu D-N. 2019. The characteristics and phylogenetic relationship of two complete mitochondrial genomes of Matrona basilaris (Odonata: Zygoptera: Calopterygidae). Mitochondrial DNA B. 4(1):1745–1747.
- Lin C-P, Chen M-Y, Huang J-P. 2010. The complete mitochondrial genome and phylogenomics of a damselfly, Euphaea formosa support a basal Odonata within the Pterygota. Gene. 468(1–2):20–29.
- Lorenzo-Carballa MO, Thompson DJ, Cordero-Rivera A, Watts PC. 2014. Next generation sequencing yields the complete mitochondrial genome of the scarce blue-tailed damselfly, Ischnura pumilio. Mitochondrial DNA. 25(4):247–248.
- Nguyen L-T, Schmidt HA, Von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Okude G, Fukatsu T, Futahashi R. 2020. Interspecific crossing between blue‐tailed damselflies Ischnura elegans and I. senegalensis in the laboratory. Entomol. Sci. 23:165–172.
- Okuyama H, Takahashi J. 2017. The complete mitochondrial genome of Mnais costalis Selys (Odonata: Calopterygidae) assembled from next generation sequencing data. TOMBO. 59:77–83.
- Posada D, Crandall KA. 1998. MODELTEST: testing the model of DNA substitution. Bioinformatics. 14(9):817–818.
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542.
- Shao H, Li Q, Liu Y. 2021. The complete mitochondrial genome of Ceriagrion fallax (Odonata: Zygoptera: Coenagrionidae) and phylogenetic analysis. Mitochondrial DNA B Resour. 6(2):491–492.
- Song N, Li X, Yin X, Li X, Yin J, Pan P. 2019. The mitochondrial genomes of palaeopteran insects and insights into the early insect relationships. Sci Rep. 9(1):17765.
- Wang L-J, Lin M-Y, Shiao S-F, Sung C-H. 2019. The complete mitochondrial genome of Psolodesmus mandarinus McLachlan, 1870 (Odonata: Calopterygidae). Mitochondrial DNA B. 4(1):337–339.
- Xu S, Guan Z, Huang Q, Xu L, Vierstraete A, Dumont HJ, Lin Q. 2018. The mitochondrial genome of Atrocalopteryx melli Ris, 1912 (Zygoptera: Calopterygidae) via Ion Torrent PGM NGS sequencing. Mitochondrial DNA B Resour. 3(1):115–117.
- Yu P, Cheng X, Ma Y, Yu D, Zhang J. 2016. The complete mitochondrial genome of Brachythemis contaminata (Odonata: Libellulidae). Mitochondrial DNA A DNA Mapp Seq Anal. 27(3):2272–2273.
- Zaka SM, Abbas N, Shad SA, Shah RM. 2014. Effect of emamectin benzoate on life history traits and relative fitness of Spodoptera litura (Lepidoptera: Noctuidae). Phytoparasitica. 42(4):493–501.
- Zhang L, Wang X-T, Wen C-L, Wang M-Y, Yang X-Z, Yuan M-L. 2017. The complete mitochondrial genome of Enallagma cyathigerum (Odonata: Coenagrionidae) and phylogenetic analysis. Mitochondrial DNA B Resour. 2(2):640–641.
